T Cell Receptor (TCR) sequencing plays a pivotal role in advancing immunology research. It allows scientists to explore the complexities of the immune system, identify novel biomarkers, and develop targeted therapies. In this article, we'll delve into TCR sequence databases—vital resources that aggregate millions of TCR sequences—and how they drive innovation in both research and clinical applications.
What is a T Cell Receptor Sequence Database?
A TCR sequence database is a specialized repository that collects, organizes, and makes accessible TCR sequences obtained from various studies and research projects. These databases provide researchers with the necessary resources to analyze and compare TCR sequences across different conditions, populations, and disease states. By aggregating vast amounts of data, TCR sequence databases facilitate in-depth studies into immune responses and offer insights into disease mechanisms.
How Do TCR Databases Impact Immunology?
TCR sequence databases contribute significantly to immunology by:
- Supporting Personalized Medicine: By understanding the diversity of TCR sequences, researchers can develop targeted therapies for cancer, autoimmune diseases, and other immune-related conditions.
- Tracking Disease Progression: TCR databases enable researchers to analyze how TCR repertoires change in response to diseases or therapies.
- Enhancing Vaccine Development: Identifying antigen-specific TCR sequences helps in designing better vaccines by targeting the right immune pathways.
Key Features of TCR Sequence Databases
TCR sequence databases are essential resources for immunologists and researchers studying T cell biology. These databases provide comprehensive access to TCR sequences, enabling the analysis of T cell diversity, specificity, and functionality in various contexts.
Extensive Data Repositories:
TCR databases like TCRdb contain vast amounts of data, with over 277 million TCR sequences from more than 8,265 samples across diverse tissues and clinical conditions. This extensive dataset allows researchers to explore TCR repertoires comprehensively (Zhang et al., 2021).
Powerful Search Functions:
Advanced search capabilities allow users to identify specific TCR sequences based on various criteria such as clinical conditions or tissue types. For instance, TCRdb offers a "fuzzy search" function that can find similar sequences even with minor discrepancies (Zhang et al., 2021).
Categorized Sample Metadata:
Databases classify samples based on metadata such as disease state or cell type. This organization facilitates comparative analyses of TCR repertoires across different conditions and enhances the understanding of immune responses in various diseases (Zhang et al., 2021).
Interactive Data Visualization:
Many databases provide interactive charts and graphs that visualize TCR diversity and gene usage patterns. For example, users can view the distribution of CDR3 lengths or V-J gene utilization through dynamic visualizations (Zhang et al., 2021).
Integration of Multiple Analysis Tools:
Tools like VisTCR allow researchers to perform comprehensive analyses of their TCR sequencing data within a user-friendly interface. This integration simplifies the process of analyzing complex datasets and enhances accessibility for researchers with varying levels of computational expertise (Qin et al., 2020).
Support for Unconventional T Cells:
Databases like UcTCRdb focus on unconventional T cell subsets and provide tools for analyzing their unique characteristics and sequence patterns. This is particularly valuable as research increasingly recognizes the importance of these unconventional populations in immune responses (Meyer et al., 2023).
Rapid Visualization Tools:
Tools such as RapTCR facilitate quick visualization and analysis of TCR repertoires by employing efficient algorithms to represent sequences in a manner that retains their biological relevance while allowing for exploratory analysis (Vandeuren et al., 2023).
Comparison of Major TCR Databases
TCR sequence databases are invaluable resources for researchers seeking to explore the complexities of the immune system. With each database offering unique features and datasets, selecting the right one depends on your research objectives. Below is an expanded comparison of the major TCR databases that are widely used in immunology research.
| Database |
Data Volume |
Key Features |
Access Link |
| TCRdb |
277 million sequences |
- Comprehensive metadata (e.g., sample information, antigen specificity)
- Powerful search functions (e.g., by gene usage, CDR3 length)
- Interactive visualization tools for TCR repertoire analysis
- Supports large-scale batch analysis |
TCRdb |
| VDJdb |
Over 100,000 TCR sequences |
- Curated antigen-specific TCR sequences
- Batch annotation tools for rapid sequence processing
- Data integration with antigen databases
- Focus on high-quality, validated sequences |
VDJdb |
| UcTCRdb |
669,900 unconventional TCRs |
- Focus on unconventional TCRs involved in unique immune responses
- User-friendly, code-free analysis tools
- Conservation analyses across species
- Detailed search options for sequence conservation and diversity |
UcTCRdb |
| TCR3D |
Structural data on TCRs |
- Geometric data on TCR–peptide–MHC interactions
- Affinity measurements for TCR binding strength
- Detailed 3D structure information
- Geometric parameters for understanding TCR specificity |
TCR3D |
1. TCRdb
TCRdb is one of the largest and most comprehensive TCR databases, containing over 277 million TCR sequences from more than 8,265 samples. These sequences are sourced from diverse conditions, such as immune responses to infections, cancer, and autoimmune diseases.

- Key Features:
- Comprehensive Metadata: TCRdb provides extensive metadata for each sequence, including sample information, antigen specificity, disease context, and more, allowing users to perform highly detailed analyses.
- Powerful Search Functions: Users can filter TCR sequences based on multiple criteria, such as gene usage, CDR3 length, and antigen specificity. This allows researchers to conduct detailed analyses of TCR diversity.
- Interactive Visualization Tools: The platform includes several interactive tools for visualizing the diversity and distribution of TCRs across different conditions. These tools help researchers assess the clonal expansion of TCRs and compare TCR repertoires in different samples.
- Batch Analysis: TCRdb supports the analysis of large datasets, allowing researchers to quickly process and compare thousands of sequences simultaneously.
TCRdb is an excellent choice for researchers looking for a comprehensive, user-friendly tool to explore TCR diversity and antigen specificity.
2. VDJdb
VDJdb is a well-known database that focuses on TCR sequences with known antigen specificity. It contains over 100,000 TCR sequences, each annotated with information about the antigen it recognizes, making it an invaluable resource for immunology research.
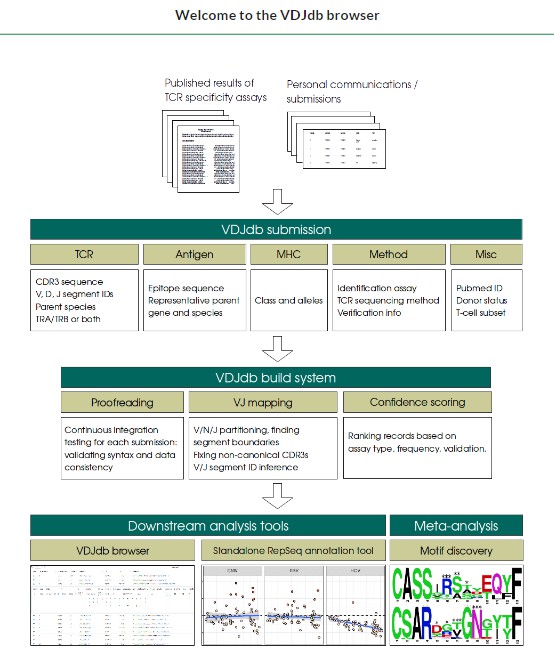
- Key Features:
- Antigen-Specific Annotations: Each sequence in VDJdb is linked to a specific antigen, allowing researchers to directly investigate how different TCR sequences recognize and respond to various pathogens, tumors, or other antigenic challenges.
- Curated Sequences: VDJdb's sequences are curated to ensure they come from high-quality, peer-reviewed studies. This ensures that researchers have access to validated TCR sequences.
- Batch Annotation Tools: The platform includes batch annotation capabilities, allowing researchers to annotate large sets of sequences quickly. This feature is particularly useful when analyzing large-scale datasets, such as those from high-throughput TCR sequencing.
- Integration with Other Databases: VDJdb integrates with several other antigen-related databases, making it easier for researchers to correlate TCR sequences with antigenic targets.
VDJdb is ideal for researchers interested in studying antigen-specific TCR sequences, particularly in the context of vaccine development, cancer immunotherapy, and autoimmune diseases.
3. UcTCRdb
UcTCRdb focuses on unconventional TCRs, which are involved in specialized immune responses. With a dataset of 669,900 unconventional TCRs, this database provides insights into immune responses that are not typically well represented in other databases.
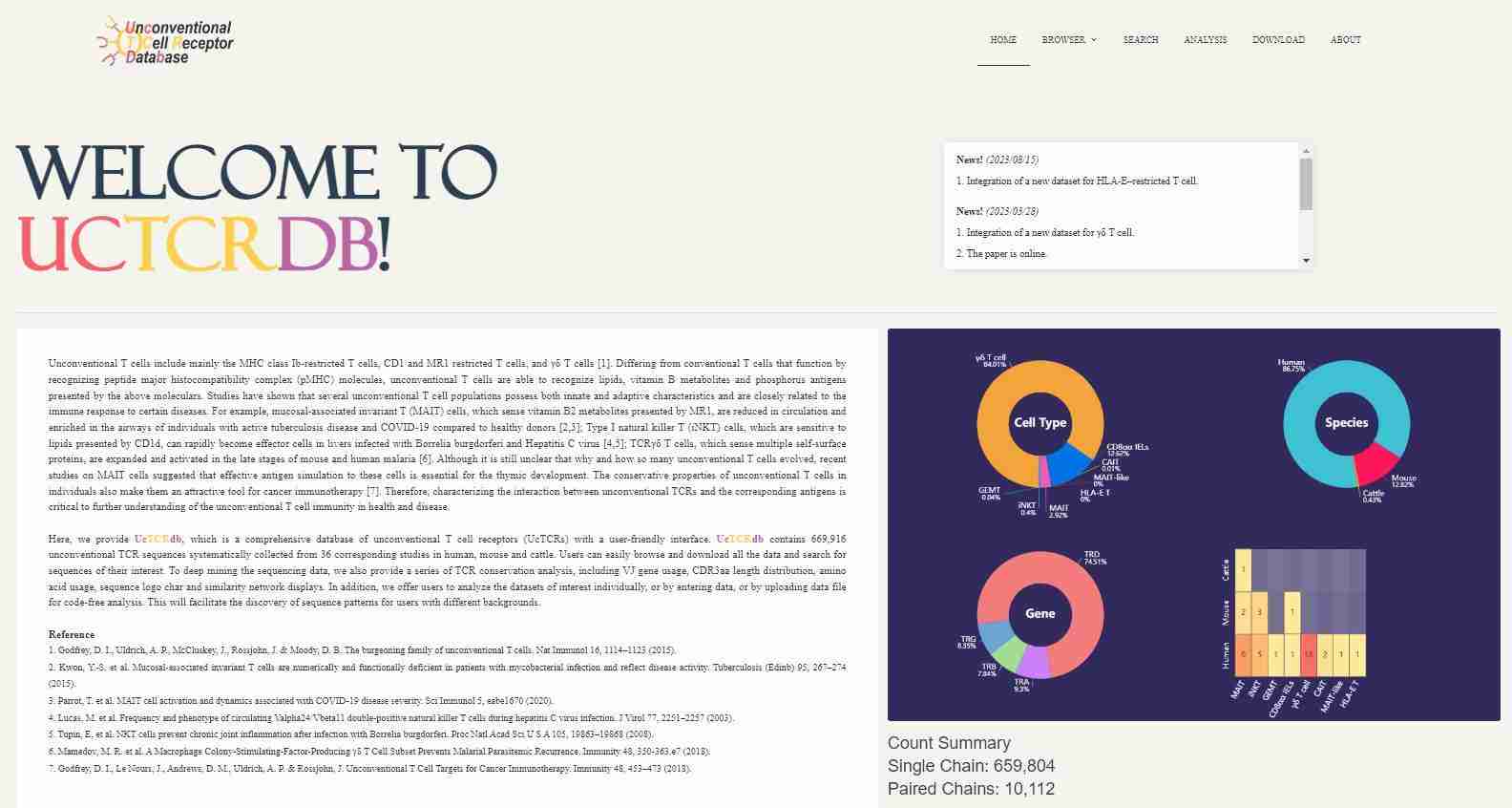
- Key Features:
- Focus on Unconventional TCRs: UcTCRdb provides data on TCRs that are involved in unique immune responses, such as those responding to non-peptide antigens or those expressed in mucosal tissues. These unconventional TCRs are essential for understanding immune functions beyond classical TCR recognition.
- Code-Free Analysis Tools: UcTCRdb features an intuitive, user-friendly interface that allows researchers to perform detailed analyses without the need for programming skills. The platform supports sequence analysis, diversity assessment, and conservation studies.
- Conservation Analyses: Researchers can explore sequence conservation across different species, enabling insights into how specific TCRs have evolved over time and their functional significance in various immune responses.
- Detailed Search Options: UcTCRdb includes advanced search functions, allowing users to filter TCR sequences based on conservation, sequence identity, and diversity.
UcTCRdb is a valuable tool for researchers studying unconventional TCRs in areas like mucosal immunity, cancer immunotherapy, and TCR-related evolutionary studies.
4. TCR3D
TCR3D is a unique database focused on the structural aspects of TCR-peptide-MHC interactions. While it doesn't store as many sequences as other databases, its focus on the three-dimensional structures of TCR interactions provides valuable insights for structural immunologists and researchers studying TCR specificity.
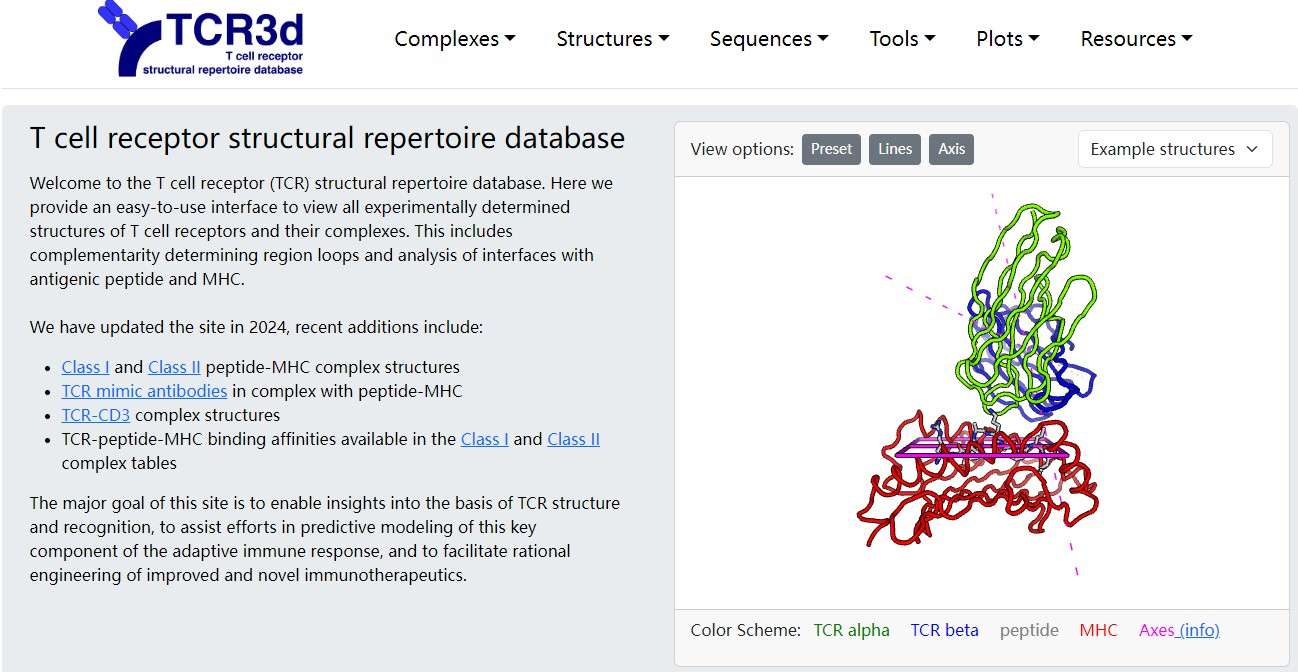
- Key Features:
- Geometric Data on TCR–Peptide–MHC Interactions: TCR3D provides detailed geometric data on the interactions between TCRs, peptides, and MHC molecules. Researchers can access information on the contact points between these molecules, as well as the geometric parameters that influence TCR binding specificity.
- Affinity Measurements: The database includes affinity measurements, providing data on how tightly different TCRs bind to their peptide-MHC complexes. This information is vital for understanding the strength of immune responses.
- 3D Structure Information: TCR3D contains 3D structural data that visualizes the shape and orientation of TCRs, peptides, and MHC molecules in complex, helping researchers better understand the mechanisms behind TCR specificity.
TCR3D is ideal for structural biologists and immunologists who are studying the molecular interactions and structural biology of TCRs.
These major TCR sequence databases provide different tools and resources to support researchers in their immunology and clinical studies. Depending on your research focus—whether it's repertoire analysis, antigen specificity, unconventional TCRs, or structural insights—you can choose the database that best fits your needs.
Challenges and Limitations in TCR Data Analysis
TCR data analysis is critical for understanding immune responses and developing immunotherapies. However, several challenges and limitations hinder the effective analysis of TCR data. Below are the key challenges, supported by relevant literature.
1. Data Complexity and Variability
The TCR repertoire is highly diverse, consisting of numerous unique sequences that can vary significantly across individuals and conditions. This complexity makes it difficult to identify meaningful patterns without sophisticated analytical tools.
A study highlights that the diversity of TCR sequences is so vast that traditional sampling methods often fail to capture the full repertoire, leading to biased estimates of diversity (Parker et al., 2015).
2. Standardization and Data Integration
There is a lack of standardization in how TCR data is recorded and shared across different studies and databases. This inconsistency complicates the comparison of datasets, making it challenging to draw meaningful conclusions from cross-study analyses.
The paper discusses the necessity for unified standards in TCR data management to facilitate better integration and comparison across studies, emphasizing that disparate formats hinder collaborative research efforts (Zhang et al., 2023).
3. Data Privacy and Access Control
Given the sensitivity of immune data, particularly when linked to patient information, privacy and access control are significant concerns. Robust data protection mechanisms are essential to ensure patient confidentiality while allowing researchers access to valuable datasets.
A report from Oak Ridge National Laboratory outlines their multi-level hierarchy for TCR data security, which includes strict access controls and compliance with cybersecurity protocols to protect sensitive information (Oak Ridge National Laboratory, 2021).
4. Limitations of Current Predictive Models
Current models for predicting TCR specificity often face limitations due to biased datasets that predominantly feature viral epitopes associated with common HLA alleles.
Research by Jiang et al. (2024) indicates that the reliance on limited epitope datasets restricts the generalization capabilities of machine learning models used for predicting TCR specificity, highlighting the need for more comprehensive training datasets.
5. Technical Limitations in Sequencing Technologies
The choice between bulk sequencing methods and single-cell sequencing technologies presents a trade-off between cost-effectiveness and detailed resolution.
According to Marktech Post (2024), bulk sequencing is high-throughput but cannot detect paired α and β chains effectively, while single-cell technologies provide this capability but are more expensive and less commonly used .
The analysis of TCR data is fraught with challenges related to complexity, standardization, privacy concerns, predictive modeling limitations, and technical constraints in sequencing technologies. Addressing these challenges is essential for advancing research in immunology and improving therapeutic strategies.
6. Conclusion: The Future of TCR Sequence Databases
As TCR sequencing technologies continue to evolve, the role of TCR sequence databases will only become more critical. By providing access to vast amounts of data, these databases are enabling breakthrough discoveries in immunology, cancer research, and personalized medicine. However, ongoing efforts are needed to address challenges like data standardization, privacy concerns, and better integration across platforms.
In the near future, we can expect even more advanced search functions, visualization tools, and data analytics capabilities to emerge, further improving the usability and value of TCR sequence databases.
Get Started with TCR Sequence Analysis Today!
Are you ready to dive deeper into TCR sequencing and analysis? CD Genomics offers comprehensive TCR sequencing services to help you explore the intricacies of TCR repertoires and unlock new insights in immunology. Whether you're studying cancer, autoimmune diseases, or vaccine development, our TCR sequencing can provide the high-quality data you need.
References:
- Zhang, Y., Guo, X., Zheng, L., Zheng, C., Song, J., Zhang, Q., Kang, B., Liu, Z., Jin, L., Xing, R., Gao, R., Zhang, L., Dong, M., Hu, X., Ren, X., Kirchhoff, D., Roider H.G., Yan, T., & Zhang, Z. (2021). TCRdb: a comprehensive database for T-cell receptor sequences. Nucleic Acids Research, 49(D1), D468-D474. https://doi.org/10.1093/nar/gkaa1004
-
Greiff, V., Miho, E., & Reddy, S. T. (2019). VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Research, 48(D1), D1057-D1059. https://doi.org/10.1093/nar/gky1040
-
Meyer, K. C., Schmittgen, T. D., & Smithson, S. L. (2023). UcTCRdb: An unconventional T cell receptor sequence database for studying unconventional T cells and their immune functions. Frontiers in Immunology, 14, 1158295. https://doi.org/10.3389/fimmu.2023.1158295
- Klein, L., & O'Connor, K. (2024). TCR3D 2.0: expanding the T cell receptor structure database with new features and content. Nucleic Acids Research. https://doi.org/10.1093/nar/gkae840
-
Qin, H., Wang, Y., Liu, H., & Zhang, Z. (2020). VisTCR: An Interactive Software for T Cell Repertoire Sequencing Data Analysis. Frontiers in Genetics, 11, 771. https://doi.org/10.3389/fgene.2020.00771
-
Vandeuren, V., Meysman, P., & De Smet, F. (2023). RapTCR: Rapid exploration and visualization of T-cell receptor repertoires using novel embedding strategies. bioRxiv. https://doi.org/10.1101/2023.09.13.557604v1
-
Parker, K. R., et al. (2015). Estimating T-cell repertoire diversity: limitations of classical methods. Nature Reviews Immunology, 15(2), 103-115. https://doi.org/10.1038/nri.2014.14
-
Zhang, Y., et al. (2023). Unified cross-modality integration and analysis of T cell receptors using UniTCR. Nature Communications, 14(1), 1234. https://doi.org/10.1038/s41467-024-47461-8
-
Oak Ridge National Laboratory. (2021). TCR Data Management Plan. Retrieved from https://info.ornl.gov/sites/publications/Files/Pub154258.pdf
-
Jiang, L., Chen, H., Pinello, L., & Yuan, G.-C. (2024). Advances and challenges in predicting TCR specificity from clustering to protein language models. Nature Reviews Immunology, 24(1), 45-60. https://doi.org/10.1038/s41577-024-00850-y


 Sample Submission Guidelines
Sample Submission Guidelines




