What is a Single Nucleotide Polymorphism (SNP)?
Single nucleotide polymorphisms, commonly referred to as SNPs (pronounced "snips"), represent the prevailing form of genetic variation within the human genome. When a cell undergoes division to produce a new cell, it first duplicates its DNA to ensure each new cell inherits a complete set of genetic instructions. However, errors can occur during this replication process, akin to typographical errors, leading to alterations in the DNA sequence at specific points known as single nucleotide polymorphisms or SNPs. Each SNP denotes a variance in a single DNA building block, or nucleotide. For instance, a SNP might replace a cytosine (C) nucleotide with a thymine (T) nucleotide within a particular DNA fragment. SNPs are pervasive in human DNA, occurring roughly once every 1,000 nucleotides on average, resulting in approximately 4 to 5 million SNPs within an individual's genome. To be classified as SNPs, these variations must be present in at least 1% of the population, and scientists have identified over 600 million SNPs across global populations.
Primarily, SNPs manifest in the non-coding regions of DNA between genes. They serve as valuable biomarkers, and when located in regulatory regions within or adjacent to genes, they may directly influence gene function, potentially impacting disease susceptibility and progression.
While many SNPs exhibit negligible impact on health or development, certain genetic variations have emerged as pivotal in human health research. SNPs play a crucial role in predicting an individual's reaction to specific medications, sensitivity to environmental factors like toxins, and susceptibility to diseases. Moreover, SNPs serve as invaluable tools in tracing the transmission of genetic variants linked to diseases within familial lineages.
Cutting-edge technologies, such as high-throughput sequencing and long-read sequencing, employed by CD Genomics, facilitate the robust analysis of SNP and SNV genotyping. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
Types of SNPs
Single nucleotide polymorphisms (SNPs) are categorized based on the specific nucleotide substitutions they entail. Here are the common types of SNPs:
- Transition: Transition SNPs represent the most prevalent variation and involve substitutions within the same chemical class. This includes exchanges between purines (adenine [A] and guanine [G]) or between pyrimidines (thymine [T] and cytosine [C]). For instance, A↔G or C↔T.
- Transversion: Transversion SNPs, on the other hand, denote substitutions between purines and pyrimidines. Examples include A↔C, A↔T, G↔C, or G↔T. These substitutions occur less frequently compared to transitions due to the differing chemical properties of purines and pyrimidines.
Beyond nucleotide substitutions, SNPs can also be classified based on their genomic location or their potential impact on gene function. Some notable categories include:
- Synonymous SNPs: These SNPs reside within the coding region of a gene but do not alter the encoded amino acids. They are often referred to as silent mutations since they do not affect the resulting protein sequence.
- Non-synonymous SNPs: These SNPs occur within the coding region and lead to amino acid substitutions in the translated protein. Depending on the nature of the substitution, they can influence protein structure and function.
- Nonsense SNPs: Nonsense SNPs introduce premature stop codons within the coding sequence, resulting in truncated and usually nonfunctional proteins.
- Promoter SNPs: These SNPs reside within the promoter region of a gene, impacting the initiation of transcription and thereby influencing gene expression levels.
- Intron SNPs: Intron SNPs are situated within the non-coding regions (introns) of a gene and can influence mRNA splicing or other regulatory processes.
Understanding the various types and implications of SNPs is essential for unraveling their role in genetic diversity, disease susceptibility, and individual traits.
Recommended reading:
An Overview of SNP Genotyping Technologies.
How to Choose Suitable SNP Genotyping Method.
What Are Single Nucleotide Variants (SNVs)?
Single nucleotide variants (SNVs) represent alterations involving a single nucleotide within a DNA sequence, presenting in three primary patterns: single nucleotide substitution, single nucleotide deletion, and single nucleotide insertion. Substitution entails the mutation of one nucleotide into another, deletion involves the removal of a single nucleotide at a specific genomic location, and insertion denotes the repeated occurrence of a single nucleotide at a particular genomic site.
Within the realm of genomic variation analysis in cancer, a single nucleotide variant distinctive to cancer cells compared to normal tissue signifies a somatic mutation, termed SNV.
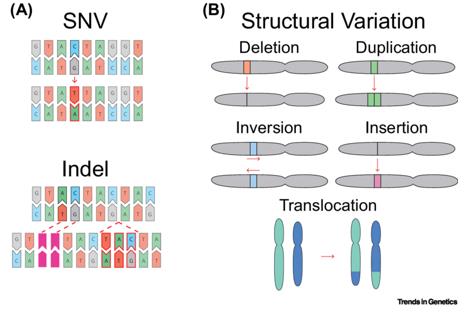 Types of Genetic Variation. (Nesta et al., 2021)
Types of Genetic Variation. (Nesta et al., 2021)
SNP vs. SNV
SNP, or single nucleotide polymorphism, refers to the substitution of a specific position in the DNA sequence with another single nucleotide (adenine, guanine, cytosine, or thymine). It stands as the most prevalent form of variation in the genome.
SNV (single nucleotide variant): This term denotes a variant at a solitary nucleotide position within the genome, irrespective of its frequency within the population. It serves as a neutral descriptor indicating a deviation from the reference sequence.
SNP (single nucleotide polymorphism): Also signifying variation at a lone nucleotide position, SNP characterizes a variation that is widespread in the population. Typically, for a variant to be termed a SNP, its prevalence in a given population generally surpasses 1%.
All SNPs are SNVs as they all denote variation at a single nucleotide. However, not all SNVs qualify as SNPs since not all single nucleotide variants are prevalent within the population.
Differences Between SNPs and CNVs
Copy number variation (CNV) refers to alterations in segments of DNA within the genome, each spanning a thousand base pairs or more. These variations can result in individuals possessing either more, fewer, or no copies of a particular gene or DNA segment compared to the typical two copies. CNVs can encompass entire genes or larger genomic regions, potentially impacting gene dosage and function.
SNPs (single nucleotide polymorphisms) and CNVs represent distinct forms of genetic diversity, yet both wield significant influence over an individual's genotype and potential phenotype. When investigating the genetic underpinnings of certain diseases or traits, researchers often explore the relationship between these two variations. Integrating insights from both SNPs and CNVs enables a more comprehensive understanding of genetic factors.
In genomic association studies, SNP arrays serve as invaluable tools, capable of detecting not only SNPs but also CNVs. By leveraging these arrays, researchers can simultaneously assess the impact of both types of genetic variants within the same study cohort. This integrated approach enhances the depth and precision of genetic investigations, shedding light on the intricate interplay between SNPs and CNVs in shaping biological traits and disease susceptibility.
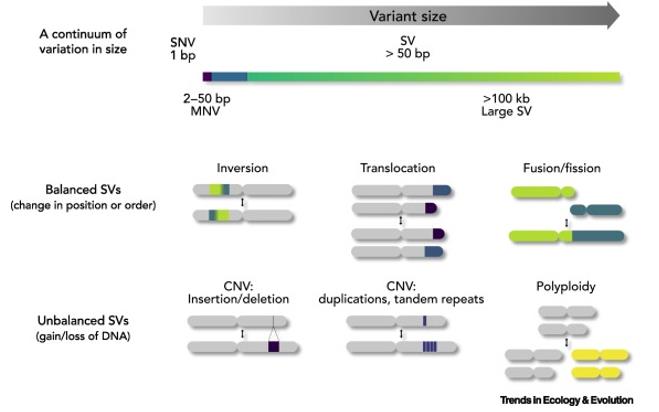 SNP and CNV. (Mérot et al., 2020)
SNP and CNV. (Mérot et al., 2020)
The Importance of SNP and SNV Genotyping
The importance of SNP and SNV genotyping lies in its ability to unravel the intricate genetic landscape underlying traits, diseases, and individual variations.
- Disease Susceptibility: Many diseases, including complex disorders like cancer and diabetes, are influenced by genetic variations. SNP genotyping enable researchers to identify specific genetic markers associated with disease susceptibility. This knowledge aids in risk assessment, early detection, and the development of targeted treatments.
- Pharmacogenomics: Individual responses to drugs can vary significantly due to genetic differences. SNP and SNV genotyping help predict how individuals will metabolize drugs, their likelihood of experiencing adverse effects, and their responsiveness to particular medications. This personalized approach to medicine, known as pharmacogenomics, enhances treatment efficacy and minimizes adverse reactions.
- Population Studies: SNV and SNP genotyping facilitate large-scale population studies aimed at understanding genetic diversity, ancestry, and evolutionary history. By analyzing genetic variations across diverse populations, researchers gain insights into migration patterns, population admixture, and genetic predispositions to certain traits or diseases.
- Precision Medicine: The era of precision medicine emphasizes tailoring healthcare interventions to individual genetic profiles. SNP genotyping plays a pivotal role in this paradigm by identifying genetic markers associated with specific diseases or treatment responses. By incorporating genetic information into clinical decision-making, precision medicine optimizes therapeutic outcomes and minimizes adverse effects.
- Biomarker Discovery: SNPs and SNVs serve as valuable biomarkers for disease diagnosis, prognosis, and treatment monitoring. By identifying genetic variations associated with disease progression or treatment response, researchers can develop biomarker-based assays for early detection, disease monitoring, and therapeutic efficacy assessment.
- Functional Genomics: SNP and SNV genotyping contribute to functional genomics studies aimed at understanding how genetic variations influence gene expression, protein function, and cellular pathways. By correlating genotype with phenotype, researchers elucidate the molecular mechanisms underlying disease pathogenesis and identify potential therapeutic targets.
Comparing SNP and SNV Genotyping Techniques
- PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism)
PCR-RFLP stands as a classic method for SNP detection. Initially, the target fragment undergoes amplification via PCR, followed by digestion of the PCR product utilizing a specific restriction endonuclease. Should a SNP be present and alter the endonuclease cleavage site, the length of the digested fragment undergoes modification. These alterations are discernible through electrophoresis, allowing for the identification of SNP variations.
The TaqMan approach employs a distinct fluorescently labeled probe to discern SNPs. During PCR, if the target SNP site perfectly matches the probe, disassembly occurs, releasing fluorescence. The intensity of this fluorescence serves as an indicator for determining the SNP species, offering a sensitive and accurate genotyping solution.
Advancements in sequencing technology have enabled the simultaneous detection of thousands of SNPs. Deep sequencing of either the entire genome or specific gene regions facilitates the identification of a vast number of SNPs, providing comprehensive insights into genetic variations and their implications.
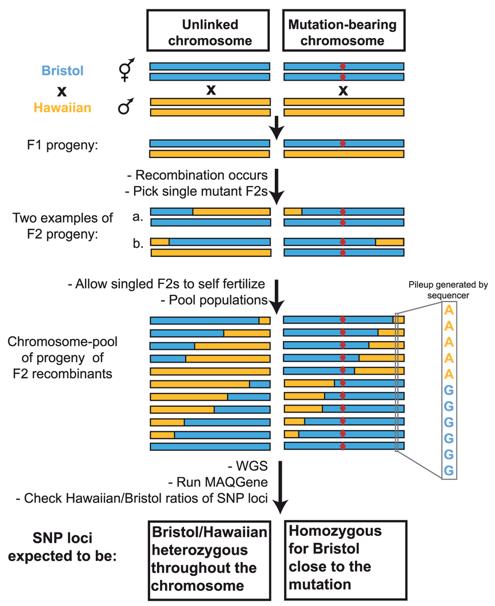 Principle of the WGS-SNP strategy. (Doitsidou et al., 2010)
Principle of the WGS-SNP strategy. (Doitsidou et al., 2010)
Gene microarray represent another high-throughput method capable of detecting millions of SNPs concurrently. The sample's DNA undergoes fragmentation and hybridization with pre-designed probes. By assessing the signal intensity post-hybridization, the type of SNP can be determined swiftly and efficiently.
Recommended reading: The Applications of SNP Microarray.
Sanger sequencing, a traditional DNA sequencing technique, remains a reliable method for SNP detection. Initially, the target region undergoes PCR amplification before sequencing via the Sanger method. By comparing the sequencing results with a reference sequence, SNPs can be accurately identified, providing valuable genetic information.
In summary, various genotyping techniques offer diverse approaches for detecting SNPs and SNVs, each possessing distinct advantages in terms of sensitivity, throughput, and accuracy. These methods play a pivotal role in unraveling the complexities of genetic variations, thereby advancing our understanding of genetics and genomics.
References:
- Doitsidou, Maria, et al. "C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy." PloS one 5.11 (2010): e15435.
- Mérot, Claire, et al. "A roadmap for understanding the evolutionary significance of structural genomic variation." Trends in Ecology & Evolution 35.7 (2020): 561-572.
- Nesta, Alex V., Denisse Tafur, and Christine R. Beck. "Hotspots of human mutation." Trends in Genetics 37.8 (2021): 717-729.


 Sample Submission Guidelines
Sample Submission Guidelines
 Types of Genetic Variation. (Nesta et al., 2021)
Types of Genetic Variation. (Nesta et al., 2021) SNP and CNV. (Mérot et al., 2020)
SNP and CNV. (Mérot et al., 2020) Principle of the WGS-SNP strategy. (Doitsidou et al., 2010)
Principle of the WGS-SNP strategy. (Doitsidou et al., 2010)