Single nucleotide polymorphism (SNP) analysis has been rapidly evolving since the launch of the Human Genome Project (HapMap Program) in 1990. SNP refers to the difference in a single nucleotide and is the most common type of genetic variations among individuals. On average, approximately 1,000 nucleotides in the human genome may result in a single nucleotide polymorphism, some of which may be associated with disease. It has been widely used in the fields of assisted reproduction, hematopoietic tumor and animal breeding.
Services you may interested in
What are the Applications of SNP Chips in the Field of Assisted Reproduction
Karyotyping in infertile couples Chromosomal abnormality is an important factor leading to infertility or a history of adverse pregnancy. Chromosomal abnormalities account for 3.3-5% of infertile patients. And the proportion of chromosomal abnormalities is as high as 10% in couples who have a history of adverse pregnancy. Traditional karyotyping methods, such as Giemsa banding and spectral karyotype analysis, have a low resolution of 0.1-5Mb, which can only detect chromosomal structural abnormalities involving large segments. The SNP microarray can be used to analyze the karyotype with higher resolution, which can detect small microdeletions and duplications. In addition, SNP microarray overcomes the challenge for traditional methods that peripheral blood lymphocyte culture is prone to failure, thus helping who are infertile or have a history of adverse pregnancy to choose a favorable mode of assisted reproductive technology. The results of in situ immunofluorescence hybridization experiments and other experimental data show that SNP microarray can accurately analyze the chimerism phenomenon of chromosome karyotype and find uniparental disomy (UPD). Novelli A et al. (2012) used high-throughput SNP microarray technology to detect the karyotype of peripheral blood lymphocytes in couples with a history of adverse pregnancy. As a result, up to 9% to 12% of genetic variants with unclear clinical significance were detected. Therefore, the occurrence of bad reproductive history can be reduced by detecting high-risk couples and providing appropriate treatment.
Preimplantation genetic diagnosis (PGD) PGD is the procedure to identify genetic defects within embryos prior to implantation. The appropriate diagnostic method is an important part of assisted reproductive technology. The SNP microarray technique has a resolution of 1.5 kb, higher than that of Array-CGH and other traditional detection methods. It can detect chromosomal unbalanced translocations, duplications and deletions of tiny fragments that cannot be detected by traditional methods. Especially for single gene diseases, SNP microarray can simultaneously detect and analyze unbalanced translocations of 23 pairs of chromosomal with abnormal number and structure. Brezina et al. (2011) used SNP microarray technique for PGD, which not only diagnosed some single gene diseases but also improved the accuracy of embryo detection, reduced the occurrence rate of adverse pregnancy and increased the pregnancy rate.
Prenatal diagnosis and prenatal screening Prenatal diagnosis and prenatal screening refer to using imaging, biochemistry, cytogenetics, and molecular biology techniques to understand the intrauterine development of the fetus before birth to diagnose congenital and genetic diseases. Becvárováet al. (2011) used SNP microarray for prenatal diagnosis of 108 fetuses. As a result, 27% of the fetuses had changes in gene copy number (copy number variants, CNVs). The results of B-ultrasound showed that 15% of the fetuses carried CNVs variation related to clinical diseases, which proved that SNP technique was very necessary in prenatal diagnosis and detection. However, the detection of high-throughput SNP requires technicians to verify whether the CNV mutation is related to clinical diseases, and about half of the CNVs mutation is a normal mutation with no clinical significance.
What are the Applications of SNP Microarray in Treatment of Hematopoietic Neoplasms
Cytogenetic analysis is essential for the diagnosis and prognosis of hematopoietic neoplasms in current clinical practice. Many hematopoietic malignancies are characterized by structural chromosomal abnormalities such as specific translocations, inversions, deletions and/or numerical abnormalities that can be identified by karyotype analysis or fluorescence in situ hybridization (FISH) studies. Single nucleotide polymorphism (SNP) arrays offer high-resolution identification of copy number variants (CNVs) and acquire copy-neutral loss of heterozygosity (LOH)/UPD that are usually not identifiable by conventional cytogenetic analysis and FISH studies. As a result, SNP arrays have been increasingly applied to hematopoietic neoplasms to search for clinically-significant genetic abnormalities. A large number of CNVs and UPDs have been identified in a variety of hematopoietic neoplasms. CNVs detected by SNP microarray in some hematopoietic neoplasms are of prognostic significance. A few specific genes in the affected regions have been implicated in the pathogenesis and may be the targets for specific therapeutic agents in the future. A proposed flow chart for the application of SNP microarray in hematopoietic malignancies is presented in Figure 1.
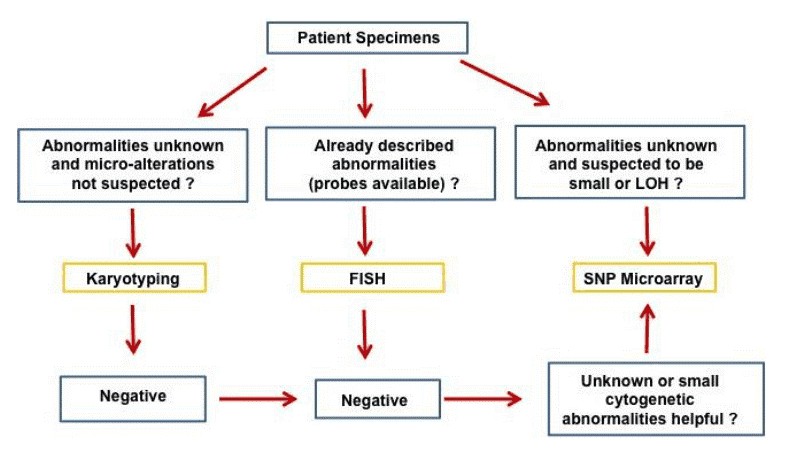 Figure 1. Proposed application of SNP array in detecting and studying hematopoietic malignancies (Song J, et al.2016)
Figure 1. Proposed application of SNP array in detecting and studying hematopoietic malignancies (Song J, et al.2016)
What are the Applications of SNP Microarray in Animal Breeding and Selection
The genetic markers associated with economic traits have been widely explored for animal breeding. Among these markers, SNPs are gradually becoming a prevalent and effective evaluation tool. Since SNPs only focus on the genetic sequences of interest, it thereby reduces the required time and cost. Compared to traditional approaches, SNP genotyping techniques incorporate informative genetic background, improve the prediction accuracy of breeding and ensure high quality offspring on the farm.
Figure 2 shows the procedure for identifying SNP markers for high economic traits for selective breeding: (1) Breeders use in silico study to obtain SNP markers from a SNP markers pool or public database (e.g., Single Nucleotide Polymorphism Database, or dbSNP); (2) Breeders conduct the primary selection or pilot study to validate SNP markers from the whole genome or putative functional genes based on in silico study. The primarily selected SNP markers are estimated to be polymorphic between two distinct small germplasm pools. Usually, DNA collected from 30–35 individuals of the same line is mixed as a representative germplasm with the other representative germplasm achieved from a distinct pool of 30–35 individuals. Only polymorphic SNP markers are chosen for the next step; (3) The secondary selection is achieved by using polymorphic SNP markers and a huge population to conduct the association analysis. The SNP markers highly associated with the quantitative trait loci (QTL) (red star) can be used as a potential genetic marker on marker-assisted selection (MAS).
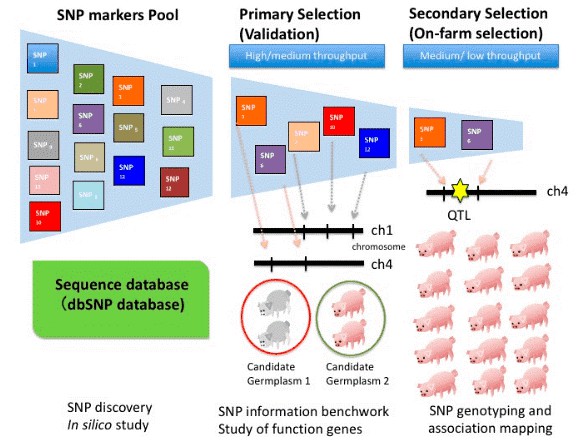 Figure 2. Procedure for identifying SNP markers with high economic traits for selective breeding. (Huang C W, et al.2015)
Figure 2. Procedure for identifying SNP markers with high economic traits for selective breeding. (Huang C W, et al.2015)
At CD Genomics, we are dedicated to providing reliable SNP genotyping services, including genotyping by sequencing, SNP microarray, MassARRAY SNP genotyping, SNaPshot Multiplex System for SNP genotyping, and TaqMan SNP genotyping.
References:
- Artini P G, Papini F, Ruggiero M, et al. Genetic screening in Italian infertile couples undergoing intrauterine insemination and in vitro fertilization techniques: a multicentric study. Gynecological Endocrinology the Official Journal of the International Society of Gynecological Endocrinology, 2011, 27(7):453-457.
- Novelli A, Grati F R, Ballarati L, et al. Microarray application in prenatal diagnosis: a position statement from the cytogenetics working group of the Italian Society of Human Genetics (SIGU), November 2011. Ultrasound Obstet Gynecol, 2012, 39(4):384-388.
- Conlin L K, Thiel B D, Bonnemann C G, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Human Molecular Genetics, 2010, 19(7):1263-1275.
- Harris, Alan R, Ferrari, et al. Genome-Wide Array-Based Copy Number Profiling in Human Placentas From Unexplained Stillbirths. Prenatal Diagnosis, 2011, 31(10):932-944.
- Brezina P R, Benner A, Rechitsky S, et al. Single-gene testing combined with single nucleotide polymorphism microarray preimplantation genetic diagnosis for aneuploidy: a novel approach in optimizing pregnancy outcome. Fertility & Sterility, 2011, 95(5):1786.e5-1786.e8.
- Becvarova V, Hynek M, Putzova M, et al. Application of SNP array method in prenatal diagnosis. Ceska Gynekol, 2011, 76(4):261-267.
- Song J, Shao H. SNP Array in Hematopoietic Neoplasms: A Review. Microarrays, 2016, 5(1):1.
- Huang C W, Lin Y T, Ding S T, et al. Efficient SNP Discovery by Combining Microarray and Lab-on-a-Chip Data for Animal Breeding and Selection. Microarrays, 2015, 4(4):570-595.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Proposed application of SNP array in detecting and studying hematopoietic malignancies (Song J, et al.2016)
Figure 1. Proposed application of SNP array in detecting and studying hematopoietic malignancies (Song J, et al.2016) Figure 2. Procedure for identifying SNP markers with high economic traits for selective breeding. (Huang C W, et al.2015)
Figure 2. Procedure for identifying SNP markers with high economic traits for selective breeding. (Huang C W, et al.2015)