SNPs are an integral part of genetic studies, and SNP genotyping is a common SNP detection method. There are many well-established SNP typing methods, each with its own characteristics and applicability.
Common typing methods include: Sanger sequencing, ligase detection reaction (LDR) typing, multiplex SNaPshot SNP genotyping, Taqman SNP genotyping, MassArray SNP genotyping and PCR-RFLP typing, etc., providing diverse SNP typing solutions for researchers.
So how to choose the right assay platform for your experiment? The article describes the technical characteristics of each typing method and compares them to help researchers.
Sanger sequencing relies on dideoxynucleotide termination reactions to generate fragments of different lengths for sequencing, and is the "gold standard" for SNP detection. The detection rate of SNP detection by Sanger sequencing is close to 100%.
The advantage of Sanger sequencing is that the results are accurate, not only to determine the type and location of mutations, but also to discover unknown SNP locus.
However, its disadvantage is low throughput and high cost. Therefore, sanger sequencing is suitable for typing scale with few locus and few samples. This typing technique is very suitable for projects with extremely high accuracy requirements or extremely small sample size (several or dozens).
PCR-RFLP
PCR-RFLP is the most classical research method for SNP typing. Its biggest advantage is that it does not require special instruments, the test operation is simple and inexpensive, and it is also suitable for the detection of medium-volume samples; the disadvantage is that many SNP locus cannot be detected by this method, and some restriction enzymes are expensive and not readily available. It is prone to false positive results and it requires a lot of manpower, takes relatively long time, and is not suitable for high-throughput large sample detection. This is an early SNP typing technology, and it is difficult to meet the needs of batch typing in terms of throughput and efficiency, so this technology is still in use only in individual laboratories.
A main disadvantage of SNP arrays is that they suffer from ascertainment bias, i.e., they cannot identify marker-trait associations in the case of SNPs that were not present in the population used for the development of the array. In addition, a typical drawback in the use of SNP arrays is the possibility that information (e.g., SNP chromosomal location) used for the design of the array is outdated and that there is no consistency in the use of SNPs among different genotyping array formats.
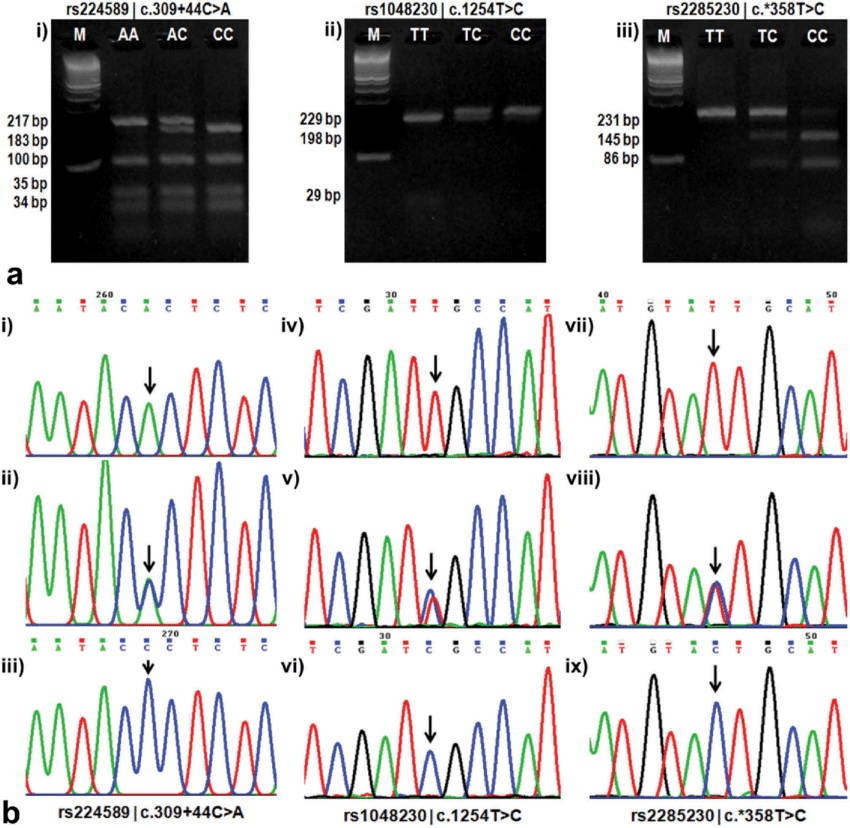 SNP genotyping by PCR-RFLP and Sanger sequencing[1]
SNP genotyping by PCR-RFLP and Sanger sequencing[1]
With the popularity of NGS technology, sequencing throughput is greatly increased and sequencing cost is significantly reduced compared to Sanger sequencing. The NGS can be used for SNP typing, not only can the sequencing read length meet the typing requirements, but also can capture hundreds of SNP locus at the same time, and the Barcode primers can distinguish different samples, allowing of high-throughput typing experiments with multiple locus and large samples in one sequencing.
Compared with the Sanger sequencing, this method not only retains the advantages of the sequencing, but also makes up for the limitation of its insufficient throughput, and greatly reduces the demand for sample volume. NGS typing is also rapidly being promoted in the field.
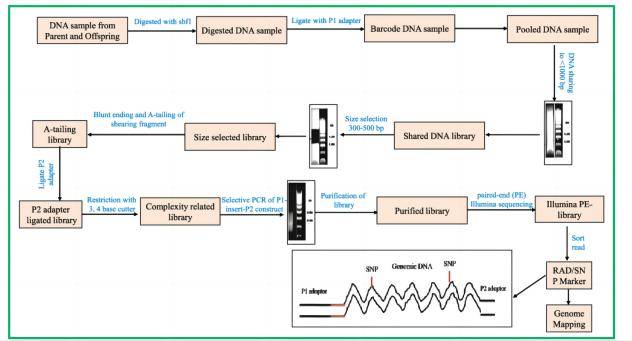 RAD-sequence: discovery and genotyping of SNPs by next-generation sequencing for genome mapping[2]
RAD-sequence: discovery and genotyping of SNPs by next-generation sequencing for genome mapping[2]
Ligase Detection Reaction (LDR)
LDR uses high-temperature ligase to realize the recognition of gene polymorphism locus. Once the high-temperature ligase detects that there is a base mismatch of gene point mutation at the junction of DNA and two complementary oligonucleotides, the ligation reaction cannot proceed.
In this method, two probes with different lengths are designed simultaneously for SNP locus. When the probe ends are unpaired with one base of the template, the ligation reaction cannot be performed and there is no ligation product; on the contrary, if the probe is completely complementary to the template DNA, the ligation reaction is therefore performed, and this specific ligation reaction can be repeated to achieve linear amplification. Finally, the detection of SNP locus is achieved by scanning the fragment length by fluorescence.
The advantage of LDR ligase detection method is that it is applicable to any polymorphic locus, the typing result is accurate, multiple reactions can be performed, the cost performance is high, and the experiment is flexible. However, compared with the TaqMan and Sanger sequencing methods, the LDR method can realize the simultaneous detection of multiple locus and improve the detection throughput, so it takes a certain amount of time to build a multiple typing scheme in the early stage. This method is very suitable for the same typing scheme and the typing requirements of continuous samples, which can increase the experimental throughput and reduce the unit price of typing.
This technology, developed by Applied Biotechnology (ABI), is a typing technology based on the principle of fluorescently labeled single base extension, and is mainly aimed at medium-throughput SNP typing projects. The reaction system contains sequencing enzymes, four fluorescently labeled ddNTPs, primers and PCR product templates, and the primers are terminated after one base extension. After detection by the ABI sequencer, the SNP locus corresponding to the extension product is determined according to the shifted peak, and the type of base incorporated can be known according to the color of the peak, thereby determining the genotype of the sample. For PCR product templates can be obtained through multiple PCR reaction systems.
SNaPshot features
(1) Accurate typing: directly obtain the base composition detection results of the locus, which are intuitive and accurate.
(2) Simultaneous detection of multiple locus: Multiple single-base detection technology can be used to detect multiple SNP locus in each reaction, while RFLP and TaqMan can only detect one at a time.
(3) Not limited by the type of SNP locus polymorphism: no matter whether the locus is heterozygous or insertion or deletion polymorphism, it can be detected in one system.
(4) Contaminated samples can be detected: If the typing peak spectrum of a sample deviates from the normal distribution, it can suggest that the sample may be contaminated or the concentration is too low, while other typing methods cannot do that. It is usually used for 10-30 SNP locus analysis.
(5) TaqMan MGB probe has a good ability to identify A/T-rich sequences.
Due to the high price of MGB probes, the Taqman probe method is usually used for the analysis of a large number of samples with a small number of SNP locus.
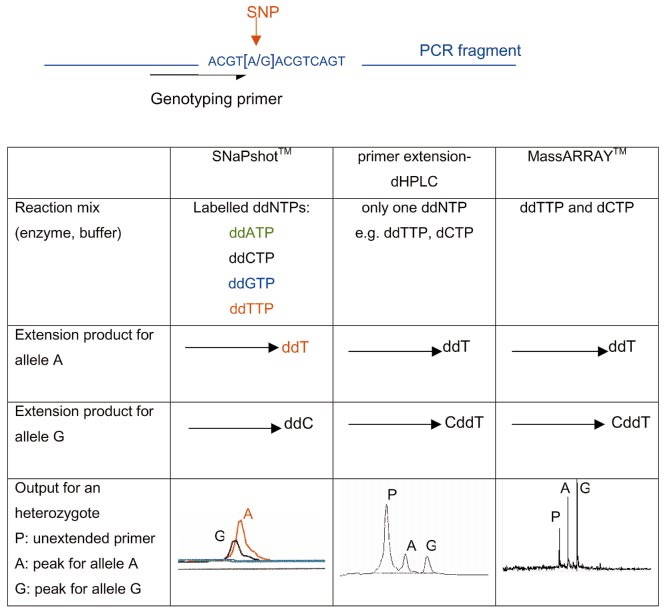 SNaPshot snp genotyping[3]
SNaPshot snp genotyping[3]
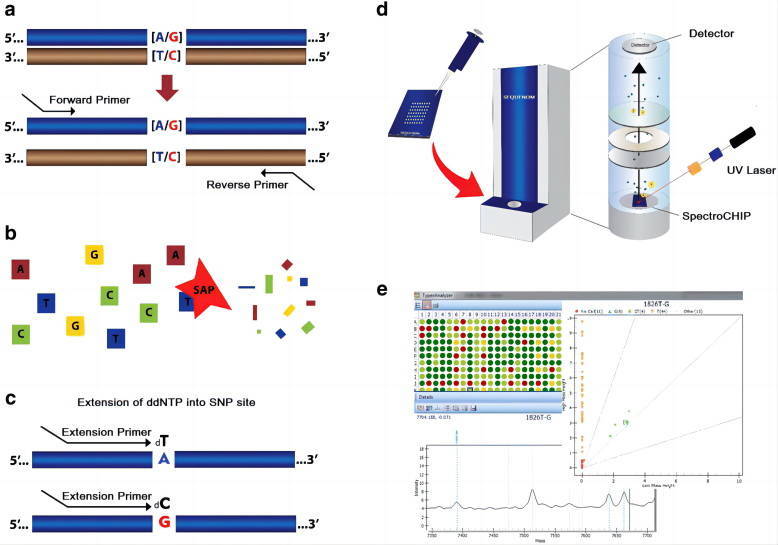
Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry, (MALDI-TOF MS) technology is the world's leading gene analysis tool launched by Sequenom.
Using the characteristic of high sensitivity to mass spectrometry,it is easy to distinguish two gene sequences containing only one different base and derive SNP typing. The iPLEX GOLD technology based on the MassARRAY platform allows the design of up to 40-weight PCR reactions and genotype detection, with flexible experimental design and high accuracy of typing results. When testing hundreds to thousands of samples for tens to hundreds of SNP locus, MassARRAY offers the best price/performance ratio and is particularly suitable for validating results found in genome-wide studies or when a limited number of study locus have been identified.
Comparison of SNP Genotyping Methods
| Genotyping method |
Technical features |
Suitable sample range |
Detection throughput |
| Sanger sequencing |
1. Simple operation and highest accuracy
2. Heavy workload and high cost |
Suitable for typing detection with small samples and large number of locus |
1,000 times/day |
| Ligase detection reaction (LDR) |
1. Applicable to any polymorphic locus
2. Accurate typing, with an accuracy of over 95%
3. High detection rate and short cycle time |
100-3000 samples, 1-20 locus |
10,000 times/day |
| Multiplex SNaPshot |
1. Accurate typing, with an accuracy of more than 95%
2. Multiple locus can be detected simultaneously
3. Not limited by the polymorphism of SNP locus
4. Not limited by the number of samples |
100-4000 samples, 8-40 locus |
10,000 times/day |
| MassARRAY |
1. High throughput: one chip can perform multiplexed assays on 384 samples
2. Up to 40 reactions per system
3. High cost performance: no fluorescent labeling, only common primers need to be synthesized
4. High sensitivity and flexibility: accuracy >95%; detection rate >90% |
Suitable for high-throughput assays with 30 to 100 locus and several thousand samples |
High throughput |
| NGS |
1. Unbiased variant discovery using only limited knowledge of the presence or nature of the variants
2. High-throughput capabilities and scalability
3. Small sample size input
4. Emerging user-friendly tools to simplify data analysis |
|
Efficiently and accurately discover and genotype thousands of SNPs |
Based on the excellent synthesis of common primers and fluorescently labeled primers, multiplex PCR amplification and sequencing technologies, CD Genomics launched SNP typing services. It covers a complete SNP typing platform from low throughput, medium throughput to high throughput, which can meet the needs of different scale projects and provide you with customized and personalized services.
References:
- Rajkumar Sankaranarayanan et al,. Protective association of A-T-T haplotype of DMT1 gene against risk of human age-related nuclear cataract March 2019 Ophthalmic Genetics 40(5):00-11
- Nazarul Hasan et al,. Recent advancements in molecular marker- assisted selection and applications in plant breeding programmes. August 2021 Journal of Genetic Engineering and Biotechnology 19(128)
- Stéphanie Le Hellard et al,. SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi automated method for data storage and analysis September 2002 Nucleic Acids Research 30(15):e74
- Maria Svidnicki et al,. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX® technologySeptember 2015 BMC Medical Genetics 16(1)
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 SNP genotyping by PCR-RFLP and Sanger sequencing[1]
SNP genotyping by PCR-RFLP and Sanger sequencing[1]
 RAD-sequence: discovery and genotyping of SNPs by next-generation sequencing for genome mapping[2]
RAD-sequence: discovery and genotyping of SNPs by next-generation sequencing for genome mapping[2] SNaPshot snp genotyping[3]
SNaPshot snp genotyping[3]
