Hepatitis B virus (HBV) is one of the most prevalent liver-associated viruses, characterized by high infectivity and a potential to cause cirrhosis and hepatocellular carcinoma (HCC). Its highly stable structure and robust immune evasion mechanisms present significant challenges for treatment. As a DNA virus, understanding its genomic structure is essential for developing effective therapies, vaccines, and diagnostic tools. This article explores the structural characteristics, replication mechanisms, genetic diversity, and clinical implications of the HBV genome.
Overview of Hepatitis B Virus
HBV represents a major global public health concern. According to the World Health Organization (WHO), approximately one-third of the global population has been exposed to HBV, with around 257 million individuals chronically infected. Chronic HBV infection significantly increases the risk of cirrhosis and HCC. HBV is a small DNA virus that primarily targets hepatocytes. Viral entry is initiated by a low-affinity interaction between the viral envelope proteins and host heparan sulfate proteoglycans (HSPG), followed by high-affinity binding to the sodium taurocholate cotransporting polypeptide (NTCP) receptor, facilitating viral invasion. Once inside the host cell, HBV undergoes translation and replication.
Service you may interested in
Resource
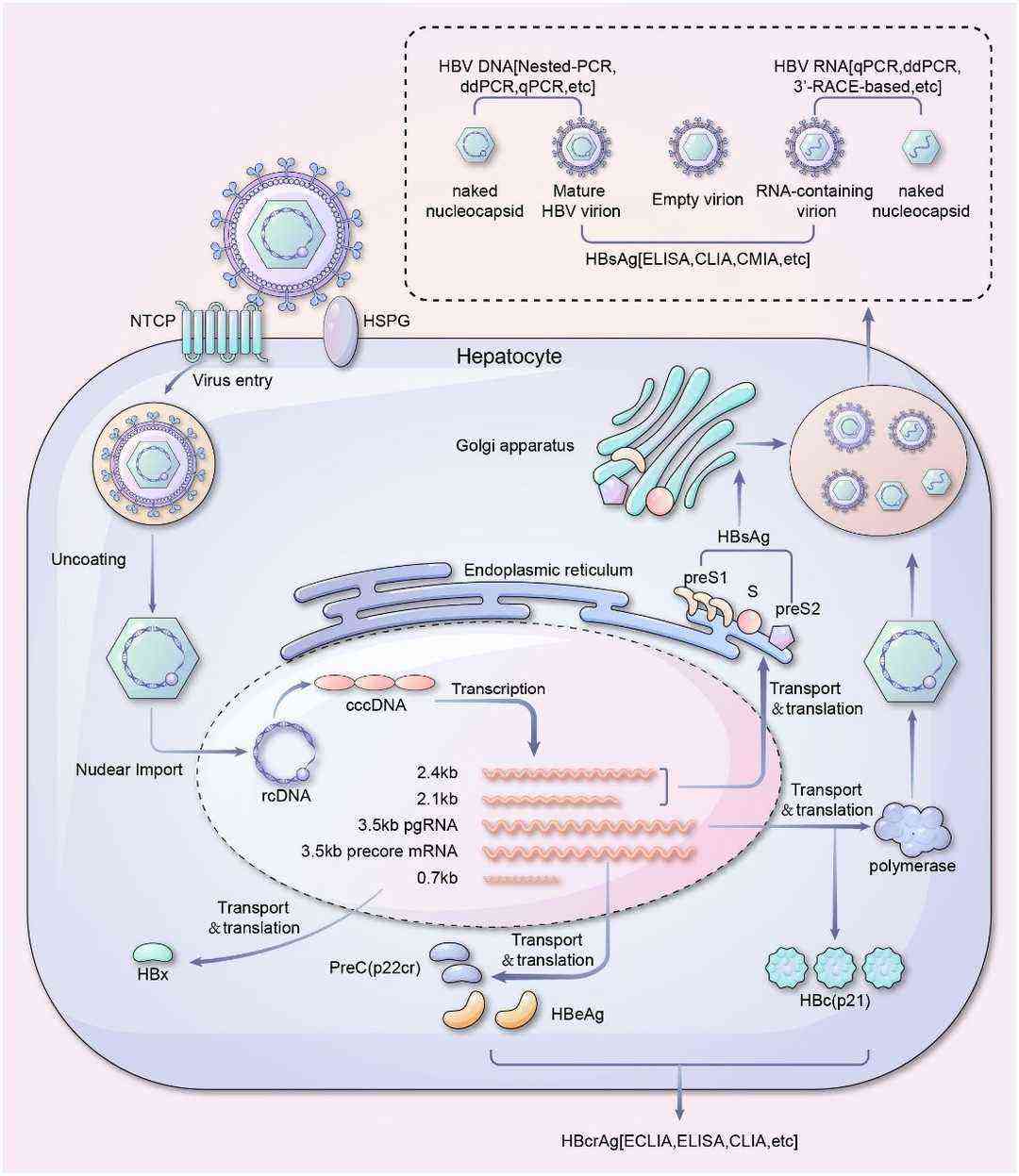 Figure 1. Life cycle and serum viral products detection of HBV.(Liu, Y.2024)
Figure 1. Life cycle and serum viral products detection of HBV.(Liu, Y.2024)
Comparison Between HBV and Hepatitis A Virus (HAV)
Unlike other hepatitis viruses, HBV is the only DNA virus that replicates via reverse transcription. In contrast, HAV is a positive-sense single-stranded RNA virus with a genome consisting of a 5' untranslated region (UTR), a single open reading frame (ORF), and a 3' UTR. HAV relies on RNA replication and does not integrate into the host genome. HAV infection induces hepatocyte death and acute hepatitis but lacks an immune evasion mechanism, allowing for complete viral clearance without chronic progression.
Structure of Hepatitis B Virus
HBV is an enveloped DNA virus with a complex structure highly adapted to the hepatic environment. It consists of an envelope, a nucleocapsid, and genetic material. The outer envelope comprises glycoproteins and lipids, incorporating hepatitis B surface antigen (HBsAg), which consists of three transmembrane glycoproteins: S, Pre-S1, and Pre-S2. These glycoproteins mediate viral entry into hepatocytes and evade host immune recognition through Pre-S1/Pre-S2 antigenic variation. Within the envelope, the nucleocapsid, primarily composed of hepatitis B core antigen (HBcAg), protects the viral genome.
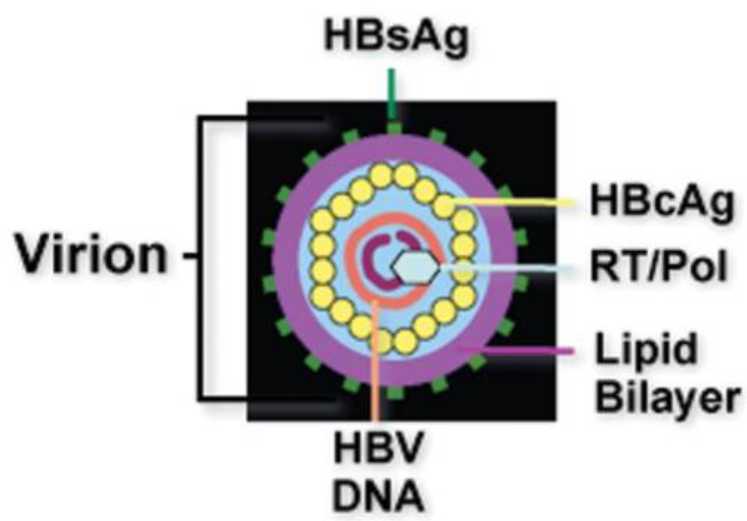 Figure 2. Schematic depiction of a hepatitis B virion.( Liang,et.al,2009)
Figure 2. Schematic depiction of a hepatitis B virion.( Liang,et.al,2009)
Genomic Composition
The HBV genome consists of approximately 3.2 kb of relaxed circular DNA (rcDNA), comprising a complete negative strand and an incomplete positive strand. It contains four overlapping ORFs: C, P, S, and X, encoding functional viral proteins:
- C: Hepatitis B core antigen (HBcAg)
- P: Polymerase (Pol)
- S: Three surface antigens (L-HBs, M-HBs, S-HBs)
- X: Hepatitis B X protein (HBx)
Upon entry into the host nucleus, rcDNA is converted into covalently closed circular DNA (cccDNA), a stable form that serves as the transcriptional template for viral mRNA and is difficult to eliminate.
HBV Replication Mechanism
HBV replication follows a unique process distinct from typical RNA and DNA viruses. After membrane fusion, the viral core enters the cytoplasm, where rcDNA is converted into plasmid-like cccDNA within the nucleus. This cccDNA acts as a template for viral gene expression and replication. Transcription from cccDNA produces various mRNAs that encode viral proteins, including surface antigens, core antigen, and polymerase. Reverse transcription by HBV polymerase generates partially double-stranded DNA, completing genome replication.
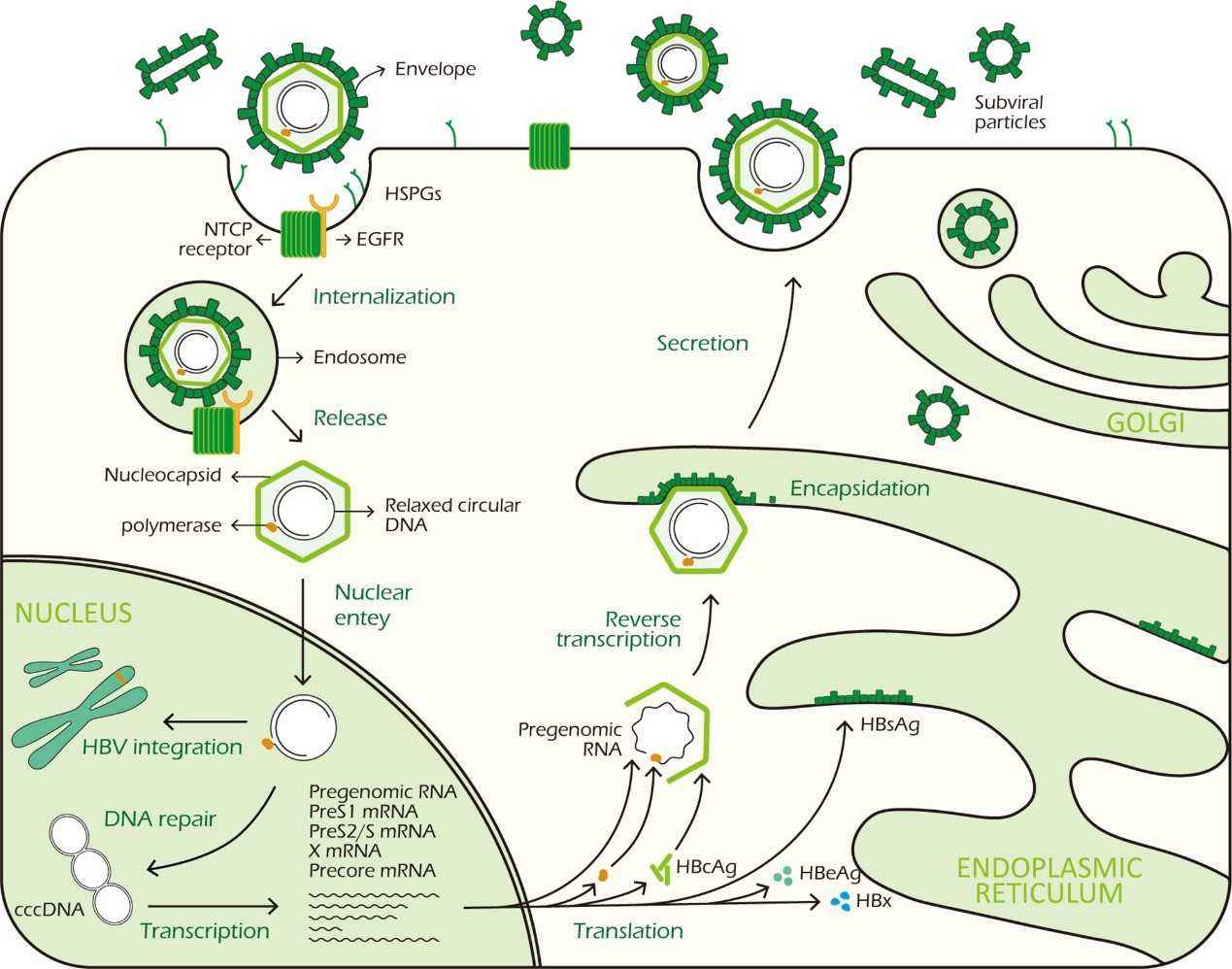 Figure3. HBV life cycle. ( Zhao,et.al,2021)
Figure3. HBV life cycle. ( Zhao,et.al,2021)
The Critical Role of cccDNA
- Essential for HBV replication: cccDNA serves as the intermediary in HBV replication and is fundamental to chronic infection persistence.
- Key factor in chronic liver disease: The stability of cccDNA in hepatocytes enables long-term viral persistence, allowing HBV reactivation under favorable conditions.
- Immune evasion mechanism: Unlike extracellular viral particles, cccDNA remains hidden within the nucleus, preventing direct immune recognition and clearance. This allows HBV to persist and contribute to chronic hepatitis progression.
Clinical Implications of HBV Genome Diversity
Genetic diversity within the HBV genome significantly impacts disease progression and treatment outcomes. For example:
- Pre-core G1896A mutation: This mutation terminates HBeAg expression (prevalence of 96% in genotype D), leading to immune evasion and an increased cirrhosis risk (5-year incidence: 38% vs. 19% for wild-type HBV).
- Basal core promoter A1762T/G1764A mutations: These suppress HBeAg transcription and increase HCC risk, with a hazard ratio of 3.3 (based on a 10-year follow-up study in Taiwan).
- Surface antigen sG145R mutation: This mutation reduces vaccine-induced neutralizing antibody efficacy by 1,000-fold. It is detected in 5.7% of mother-to-child transmission breakthrough cases in Asia.
These mutations accelerate liver fibrosis and tumor progression by altering viral protein conformations and regulatory element activities.
From a therapeutic perspective, HBV genotype characteristics and cccDNA persistence pose major challenges. The estimated half-life of cccDNA in hepatocytes ranges from 33 to 50 days, with mathematical models suggesting that complete viral eradication requires 14 years of continuous suppression. Drug resistance further complicates treatment:
- Lamivudine resistance (rtM204V mutation): The 5-year incidence of this mutation reaches 76% in untreated patients.
- Genotype C and interferon response: Genotype C exhibits a 40% lower response rate to pegylated interferon compared to genotype B.
Next-generation sequencing (NGS) enables the detection of low-frequency resistance mutations (0.1%). For patients with baseline rtM204I mutation frequencies exceeding 2%, treatment failure risk increases 4.5-fold. Therefore, integrating mutational analysis with personalized therapy is crucial for overcoming therapeutic bottlenecks.
Advances in Genetic Testing Technology
Hepatitis B virus (HBV) exhibits highly dynamic genomic evolution, posing significant challenges for clinical diagnosis and treatment. Due to the lack of proofreading function in HBV reverse transcriptase, its mutation rate reaches up to 1 × 10⁻⁴ per replication cycle, resulting in a complex viral quasispecies population within a single host. Traditional detection methods, such as Sanger sequencing, can only identify dominant mutant strains with frequencies exceeding 20%. However, next-generation sequencing (NGS) has revealed that 68% of chronic hepatitis B patients harbor low-frequency drug-resistant mutations (e.g., rtM204V/I, mutation frequency 0.1%-5%) before treatment. These cryptic variants may pose potential risks of treatment failure. For example, a 2023 longitudinal study found that patients with baseline rtA181T mutation abundance exceeding 0.3% developed multidrug resistance 8 months earlier than those without mutations (p<0.001). Additionally, the A1762T/G1764A double mutation in the core promoter region increases the risk of hepatocellular carcinoma (HCC) by 3.5 times, while Pre-S region deletion mutations are detected in 41% of HCC patients (compared to 9% in controls). These mutations promote malignant transformation by activating endoplasmic reticulum stress pathways. Therefore, clinical practice is gradually shifting towards precision management. For instance, in response to the sG145R vaccine escape mutation, neonatal immunization doses are adjusted to 10 μg; patients carrying BCP/Pre-C mutations require ultrasound combined with alpha-fetoprotein monitoring every 6 months; and patients with rtN236T mutation are contraindicated from using entecavir monotherapy. The successful implementation of these strategies depends on technologies capable of detecting variants at frequencies below 1%, highlighting the limitations of traditional methods.
Single-Molecule Real-Time Sequencing and Deep Sequencing: Decoding Viral Evolutionary Patterns
Breakthroughs in third-generation sequencing (TGS) have provided new insights into HBV evolution. Long-read sequencing technologies such as PacBio HiFi and Oxford Nanopore not only enable complete genome assembly but also track haplotype variations. A 2023 study involving 156 chronically infected individuals identified three major evolutionary patterns: 68% exhibited a "stabilized selection" pattern, where dominant strains remained stable over five years; 22% showed a "directional evolution" pattern characterized by the gradual accumulation of Pre-S2 deletions; and 10% displayed a "punctuated equilibrium" pattern, where rtS78T/sI126S dual mutations emerged under entecavir treatment pressure. Nanopore sequencing has demonstrated remarkable clinical utility, achieving 99.8% concordance with NGS in variant detection and reducing detection time from 7 days to 8 hours—crucial for rapid acute hepatitis classification. Additionally, ultra-deep sequencing of the Pre-S region (10,000× coverage) has revealed genotype-specific oncogenic mechanisms: HCC risk is 7.3 times higher in genotype C patients with Pre-S mutations compared to genotype B (95% CI 3.1–17.2), and Pre-S2 start codon mutations exhibit 89% sensitivity for early HCC detection. Notably, Pre-S1 deletions synergizing with HBx K130M mutations significantly activate the β-catenin signaling pathway (p=0.003), providing a molecular basis for targeted interventions. A 2024 multicenter trial further demonstrated that combining Pre-S mutation burden (>15%) with serum PIVKA-II testing improves HCC prediction accuracy to an AUC of 0.92 (compared to 0.78 for AFP alone).
Future Prospects: Technology Integration and Clinical Translation
Cutting-edge technologies continue to push the boundaries of detection sensitivity. CRISPR-Cas9-guided targeted enrichment has increased NGS sensitivity to 0.01%, enabling the capture of ultra-low-frequency drug-resistant mutations. Digital droplet PCR (ddPCR) allows absolute quantification of cccDNA at a single-copy level (sensitivity: 0.1 copies/cell), with results significantly correlated with fibrosis staging (r=0.71, p<0.001), serving as a key indicator for functional cure assessment. Moreover, artificial intelligence is ushering in a new era of predictive genetics: deep learning-based mutation feature analysis can predict HCC occurrence up to 3 years in advance (F1-score 0.85). However, standardization and cost control remain bottlenecks for global adoption. The 2024 World Health Organization (WHO) HBV diagnostic roadmap prioritizes the development of point-of-care third-generation sequencing devices, aiming to reduce the "sample-to-result" time to under 4 hours and improve accessibility in low- and middle-income countries.
Technology Integration: From Basic Research to Clinical Decision-Making
The synergistic application of innovative technologies is redefining HBV diagnosis and treatment paradigms. Spatial transcriptomics (e.g., 10x Genomics Visium) has revealed the regional distribution characteristics of HBV infection—viral load in liver lobule Zone 3 is 4.7 times that in Zone 1, providing a theoretical basis for localized therapy. Meanwhile, CRISPR-Dx combined with the SHERLOCK system can detect 0.1% low-frequency drug-resistant mutations within 30 minutes (specificity 99.8%), significantly enhancing clinical decision-making efficiency. Future genetic testing will extend beyond viral monitoring to integrate host immune features, epigenetic regulation, and other multidimensional data to construct personalized treatment strategies. For instance, single-cell ATAC-seq can analyze cccDNA chromatin accessibility to identify potential epigenetic therapeutic targets, while nanopore sequencing combined with AI algorithms enables real-time tracking of viral evolution for dynamic treatment adjustments.
Conclusion
The genomic characteristics of hepatitis B virus (HBV) and its persistence mechanisms present core challenges in chronic liver disease prevention and treatment. As the only DNA virus that relies on reverse transcription for replication, HBV establishes a stable viral reservoir within host hepatocytes through the long-term persistence of covalently closed circular DNA (cccDNA), allowing immune evasion and resistance to current antiviral therapies, making chronic infection difficult to eradicate.
HBV genome diversity influences viral pathogenicity and host immune responses through key site mutations, accelerating fibrosis and HCC progression. The persistence of cccDNA not only sustains viral replication but also serves as a primary bottleneck for antiviral treatment. Advances in novel detection technologies have enhanced the identification of low-frequency mutations, providing a molecular foundation for precision interventions.
Future research should focus on innovative strategies targeting cccDNA clearance, integrating multi-omics technologies with artificial intelligence models to optimize early warning systems and individualized therapies. The global implementation of rapid diagnostic technologies and the clinical translation of novel therapies are paving new paths to eliminating this public health threat. Deepening HBV genome research not only elucidates the complex virus-host interaction network but also underscores the profound value of translational science in clinical medicine.
References:
- Tsukuda S, Watashi K: Hepatitis B virus biology and life cycle. Antiviral Research 2020, 182:104925. https://doi.org/10.1016/j.antiviral.2020.104925.
- Xia Y, Guo H: Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential. Antiviral Research 2020, 180:104824. https://doi.org/10.1016/j.antiviral.2020.104824.
- Zhao F, Xie X, Tan X, Yu H, Tian M, Lv H, Qin C, Qi J, Zhu Q: The Functions of Hepatitis B Virus Encoding Proteins: Viral Persistence and Liver Pathogenesis. Frontiers in Immunology 2021, https://doi.org/10.3389/fimmu.2021.691766.
- Liu Y, Wu D, Zhang K, Ren R, Liu Y, Zhang S, Zhang X, Cheng J, Chen L, Huang J: Detection technology and clinical applications of serum viral products of hepatitis B virus infection. 2024, https://doi.org/10.3389/fcimb.2024.1402001.
- Ortonne V, Bouvier-Alias M, Vo-Quang E, Cappy P, Ingiliz P, Leroy V, Pawlotsky J-M, Chevaliez S: Performance evaluation of a fully automated deep sequencing platform for hepatitis B genotyping and resistance testing. Journal of Antimicrobial Chemotherapy 2025, https://doi.org/10.1093/jac/dkae418.
- Payne M, Ritchie G, Lawson T, Young M, Jang W, Stefanovic A, Romney MG, Matic N, Lowe CF: Evaluation of a next generation sequencing assay for Hepatitis B antiviral drug resistance on the oxford nanopore system. Journal of Clinical Virology 2025, https://doi.org/10.1016/j.jcv.2025.105762.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Life cycle and serum viral products detection of HBV.(Liu, Y.2024)
Figure 1. Life cycle and serum viral products detection of HBV.(Liu, Y.2024) Figure 2. Schematic depiction of a hepatitis B virion.( Liang,et.al,2009)
Figure 2. Schematic depiction of a hepatitis B virion.( Liang,et.al,2009) Figure3. HBV life cycle. ( Zhao,et.al,2021)
Figure3. HBV life cycle. ( Zhao,et.al,2021)