Introduction to CRISPR Off-Target Validation
The CRISPR/Cas9 system has revolutionized genetic engineering, furnishing researchers with an unparalleled tool for precise genome editing. Nonetheless, the system's propensity for off-target effects—unintended edits in non-target genomic regions—poses a pronounced concern. These off-target mutations can induce unexpected phenotypic alterations, thereby complicating the interpretation of experimental outcomes and potentially jeopardizing therapeutic applications. Addressing these challenges necessitates the implementation of stringent off-target validation protocols.
Towards this end, CD Genomics provides sophisticated CRISPR off-target validation services, employing next-generation sequencing (NGS) technologies to meticulously assess and validate the specificity of CRISPR/Cas9 edits. For the detection of potentially hundreds of off-target effects, CD Genomics utilizes multiplex PCR or probe design panels targeting these specific sites, leveraging NGS to verify the presence and frequency of edits at these loci.
How to Detect Off-Target Effects
CRISPR technology involves the formation of a complex between single guide RNA (sgRNA) and Cas proteins. The sgRNA identifies and pairs with the complementary target sequence on a DNA strand, while the Cas protein recognizes the protospacer adjacent motif (PAM) sequence. When both the PAM and target sequences match, the Cas protein cleaves the target DNA, resulting in double-strand breaks. The cell's non-homologous end joining repair mechanism then attempts to fix the break, often leading to errors that facilitate gene editing.
However, a significant challenge of CRISPR technology is off-target effects, where the sgRNA may mistakenly bind to non-target sequences, leading to unintended edits in other genomic regions. This raises concerns about the verification of gene functions and the safety of gene editing applications. Therefore, off-target detection is crucial, with both first-generation and next-generation sequencing methods available for this purpose.
For a detailed overview of CRISPR off-target detection methods, you can refer to the articles
- Next Generation Sequencing Validating Your CRISPR/Cas9 Edit
- Summary of CRISPR-Cas9 off-target Detection Methods
Advantages of CRISPR Off-Target Validation Service
- Multiplex Analysis: Analyze hundreds target sites simultaneously within a single experiment, improving efficiency and providing deeper insights into the effects of CRISPR editing.
- Customizable Design: Leverage our advanced design tool to create tailored primers that ensure precise amplification of both on-target and off-target sequences, meeting your specific research needs with accuracy.
- High Sensitivity and Specificity: CD Genomics utilizes NGS-based platforms to achieve deep sequencing coverage, enabling the detection of low-frequency off-target mutations that might otherwise go unnoticed. This high sensitivity is essential for ensuring that gene editing is precise and effective.
- Comprehensive Reporting: The validation process includes detailed reports that provide insights into off-target effects, enabling researchers to make informed decisions regarding their CRISPR designs and applications. CD Genomics' experts offer consultation to interpret results and suggest modifications to improve specificity.
- Streamlined Workflow: The end-to-end validation process simplifies the analysis of CRISPR edits. By offering a one-stop service from sample preparation to final reporting, CD Genomics enhances efficiency, allowing researchers to focus on their primary objectives without being bogged down by complex validation procedures.
Applications of CRISPR Off-Target Validation
- Gene Therapy: CRISPR edits must be precise in therapeutic contexts to ensure patient safety by preventing off-target mutations. Rigorous off-target validation protocols are essential to confirm that genome modifications are confined to intended sites, minimizing potential adverse effects.
- Agricultural Biotechnology: CRISPR technology is transforming agricultural biotechnology, allowing for the creation of genetically modified crops. Off-target validation is critical to ensure that desired traits are integrated into the crop genome without adversely affecting plant health or development, thereby ensuring the safety and sustainability of agricultural products.
- Basic Research: In fundamental research, precise genetic editing is necessary to understand gene function. Reliable off-target validation allows researchers to accurately interpret results, ensuring that observed phenotypes are due only to intended modifications, which is vital for advancing knowledge of genetic pathways and mechanisms.
CRISPR Off-Target Validation Workflow
The CRISPR off-target validation workflow at CD Genomics involves submitting samples, constructing libraries of target regions, conducting high-throughput sequencing, performing bioinformatics analysis for reference alignment and visual annotation, assessing target activity etc.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
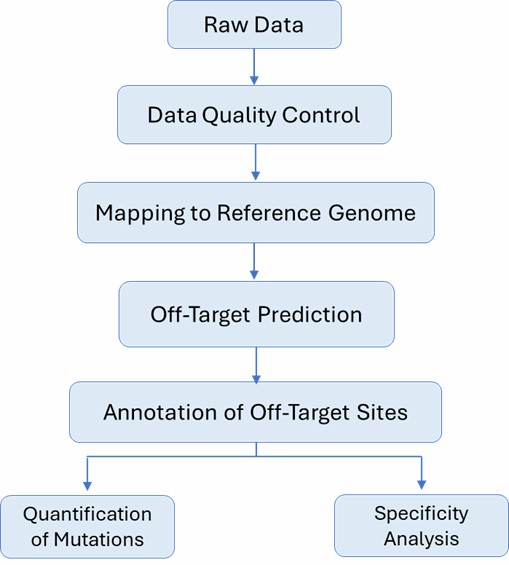
Genome Editing with CRISPR: How to Effectively Minimize Off-Target Effects
Deliverables
- Raw data
- Experimental results
- Data analysis report
References
- Park S H, Lee C M, Bao G. Identification and Validation of CRISPR/Cas9 Off-Target Activity in Hematopoietic Stem and Progenitor Cells//Stem Cell Assays: Methods and Protocols. New York, NY: Springer US, 2022: 281-306.
- Kang S H, Lee W, An J H, et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nature communications, 2020, 11(1): 3596.
Partial results are shown below:
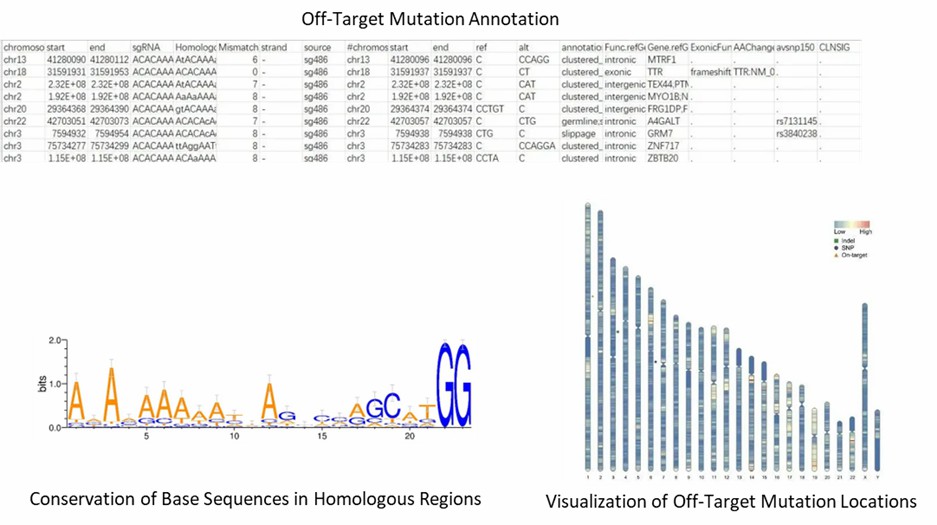
1. How can I minimize off-target effects when designing my CRISPR experiments?
To reduce off-target effects, ensure that your gRNA has optimal GC content (40%-60%) and is approximately 17 nucleotides in length. Utilize computational tools to predict off-target sites and choose gRNAs with fewer potential matches in the genome.
2. How does CD Genomics ensure the accuracy of its off-target validation services?
CD Genomics employs state-of-the-art NGS technologies coupled with rigorous bioinformatics analyses to ensure high sensitivity and specificity in detecting off-target mutations. Comprehensive reporting and expert consultation further enhance the reliability of our validation services.
3. Is off-target validation necessary for all CRISPR experiments?
While off-target validation is especially critical for therapeutic applications and projects with high stakes, it is also advisable in research settings where precision is paramount. This validation helps ensure that the results are attributable to intended modifications.
Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment
Journal: Nature Communications
Impact factor: 16.6
Published: 17 July 2020
Background
The CRISPR–Cas system allows precise gene editing, but can cause off-target mutations, especially in large genomes. While NGS methods are used to detect these mutations, they often lack sensitivity. A new method using CRISPR endonucleases and PCR enhances the detection of low-abundance off-target mutations, improving accuracy for safe gene therapies.
Materials & Methods
Sample Preparation
- Escherichia coli BL21 (DE3)
- DNA extraction
Method
- PCR
- NGS
- Targeted deep sequencing
- Off-target mutation analysis
- Mutation frequency calculation
Results
To assess off-target mutations with CRISPR–Cas9, CRISPR amplification was used to detect predicted off-target mutations in HEK293FT cells. This method, enhanced by Cas12a for selective amplification, revealed significant increases in indels and allowed detection of low-frequency mutations that conventional NGS missed. CRISPR amplification also distinguished true off-target effects from false positives, proving effective for sensitive screening.
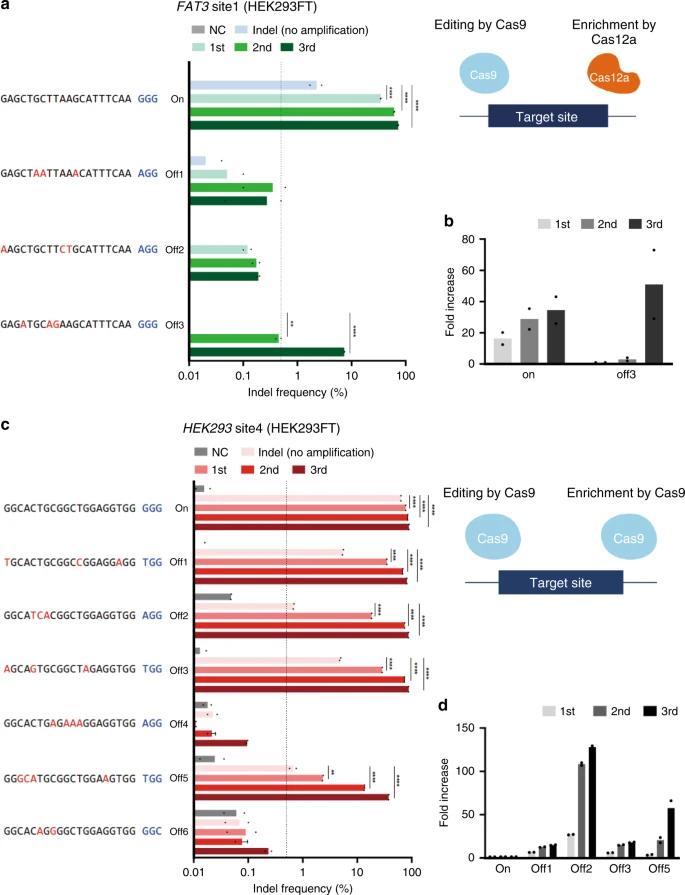 Fig. 1: Detection of off-target mutations induced by CRISPR–Cas9.
Fig. 1: Detection of off-target mutations induced by CRISPR–Cas9.
CRISPR amplification effectively detected single-base A>G substitutions induced by adenine base editors (ABE) in HEK293FT cells, revealing mutations that conventional NGS missed and showing high sensitivity for identifying specific off-target changes.
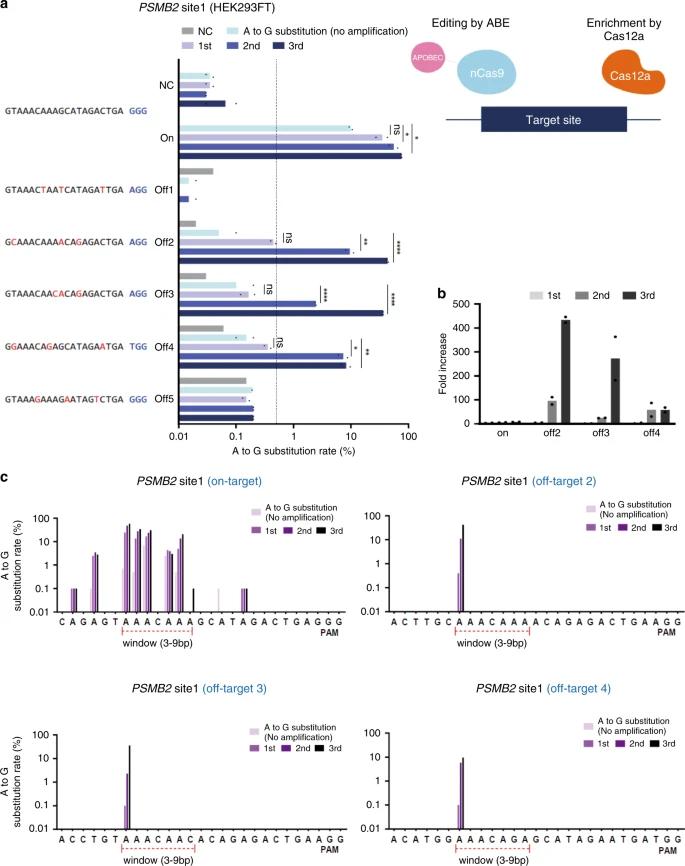 Fig. 2: Detection of single-base off-target mutations induced by ABE.
Fig. 2: Detection of single-base off-target mutations induced by ABE.
Conclusion
The authors developed a CRISPR amplification method to improve detection of off-target mutations by enriching mutant DNA and eliminating wild-type DNA. This approach enhances sensitivity, revealing low-abundance mutations missed by conventional NGS. It is effective for various CRISPR effectors, including Cas9, Cas12a, and adenine base editors (ABE), and provides accurate analysis for gene therapy and precision genome engineering.
Reference
- Kang S H, Lee W, An J H, et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nature communications, 2020, 11(1): 3596.
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
