Where is the AAV Genome Integration Site
Adeno-Associated Virus (AAV) vectors are known for their ability to transduce transgenes efficiently into various somatic cells within patients' organs. Although traditionally considered episomal vectors, AAV genomes can integrate into the host cell's genome, with integration frequencies ranging from 0.1% to 3% in various cell types such as hepatocytes, fibroblasts, muscle cells, and neurons. Integration hotspots have been identified at genomic sites characterized by palindromic sequences, CpG islands, transcriptional start sites (TSSs), and active genes.
One significant integration site is the AAVS1 locus, which is located within the PPP1R12C gene on chromosome 19 of the human genome. This site is recognized for its capacity to accommodate the insertion of exogenous genes while maintaining stable gene expression without adversely affecting normal cellular function and development. Consequently, AAVS1 is often referred to as a "safe harbor" site, making it an ideal target for gene editing and other applications.
How does AAV Integrate
The integration of AAV into the host genome is a complex process that relies heavily on host factors, as AAV vectors do not encode proteins necessary for integration. AAV integration typically occurs at sites of double-strand breaks (DSBs) or nicks in the host genome. This integration can occur through homologous recombination or non-homologous end joining mechanisms. The presence of free chromosomal ends available for ligation with AAV genomes suggests that nuclear proteins involved in the DNA damage response play a critical role in regulating the integration process.
While preclinical and clinical studies have indicated a favorable safety profile for AAV integrations, concerns remain regarding their long-term genotoxic potential. High doses of AAV in preclinical models have been associated with hepatocellular carcinoma and clonal expansions due to the activation of oncogenes. Consequently, while AAV integration ensures sustained transgene expression and potential other benefits, it also necessitates careful consideration of safety measures.
Introduction to AAV Integration Site Analysis
AAV Integration Site Analysis serves as a vital tool for understanding the genomic behavior of AAV vectors post-administration. This analysis identifies and characterizes AAV integration sites within the host genome, providing insights into the implications for gene therapy applications. CD Genomics employs advanced methods, including PCR and next-generation sequencing (NGS) platforms like Illumina and PacBio, to facilitate high-throughput sequencing and accurate mapping of integration sites.
One promising method employed in this analysis is hybrid-capture-based target enrichment sequencing (TES). This approach utilizes specially designed baits to isolate regions of interest (ROI), such as long terminal repeats (LTR) and transgenes, from a traditional whole genome sequencing (WGS) library. Notably, the TES method does not necessitate intact genomic ends, making it particularly effective for detecting truncated insertions that may arise from AAV therapies. By leveraging TES, researchers can gain deeper insights into the integration patterns of AAV vectors, which is vital for optimizing gene therapy strategies.
Custom bioinformatics pipelines are also used to analyze sequencing data, effectively identifying integration loci and vector rearrangements.
Advantages of AAV Integration Site Analysis Service
- Comprehensive Characterization: CD Genomics' AAV Integration Site Analysis service enables the detailed characterization of AAV integration sites, which is essential for understanding the long-term effects of AAV vectors on the host genome.
- Enhanced Safety Assessment: By identifying integration hotspots and potential oncogenic insertions, our service helps mitigate risks associated with insertional mutagenesis and ensures safety.
- Optimized Strategies: Insights from AAV integration site analyses allow for the refinement of approaches, facilitating targeted delivery and minimizing adverse effects associated with random integration.
- Support for Regulatory Compliance: Comprehensive integration site analysis is increasingly required for regulatory submissions. Our services assist clients in meeting the necessary regulatory standards.
Applications of AAV Integration Site Analysis
The AAV Integration Site Analysis service offered by CD Genomics has broad applications across various fields, including:
- Oncology Research: The analysis of integration sites is critical in oncology research, particularly in assessing the genotoxicity of AAV vectors and their potential for tumorigenesis.
- Immunology Studies: AAV integration can be utilized for benefits in immunological applications, such as enhancing T-cell functionality or addressing immunodeficiencies.
- Animal Models: Integration site analysis in preclinical models provides valuable data on the potential effects of AAV administration and assists in predicting outcomes.
AAV Integration Site Analysis Workflow
The AAV Integration Site Analysis workflow at CD Genomics involves several key steps: first, tissues or cell samples from AAV-treated subjects are collected for genomic analysis. High-quality genomic DNA is extracted and prepared into libraries using a sonication-based linker-mediated PCR method (SLiM). These libraries are then sequenced using hybrid-capture-based target enrichment sequencing methods. The sequencing data undergo rigorous analysis using the RAAVioli pipeline to identify and characterize integration sites, which are subsequently interpreted and mapped to genomic features to provide insights into safety and efficacy.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- Raw data
- Experimental results
- Data analysis report
References
- Dalwadi D A, Calabria A, Tiyaboonchai A, et al. AAV integration in human hepatocytes. Molecular Therapy, 2021, 29(10): 2898-2909.
- Greig J A, Martins K M, Breton C, et al. Integrated vector genomes may contribute to long-term expression in primate liver after AAV administration. Nature Biotechnology, 2023: 1-11.
- Henckaerts E, Dutheil N, Zeltner N, et al. Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proceedings of the National Academy of Sciences, 2009, 106(18): 7571-7576.
- Dyall J, Szabo P, Berns K I. Adeno-associated virus (AAV) site-specific integration: Formation of AAV–AAVS1 junctions in an in vitro system. Proceedings of the National Academy of Sciences, 1999, 96(22): 12849-12854.
- Calabria A, Cipriani C, Spinozzi G, et al. Intrathymic AAV delivery results in therapeutic site-specific integration at TCR loci in mice. Blood, The Journal of the American Society of Hematology, 2023, 141(19): 2316-2329.
Partial results are shown below:
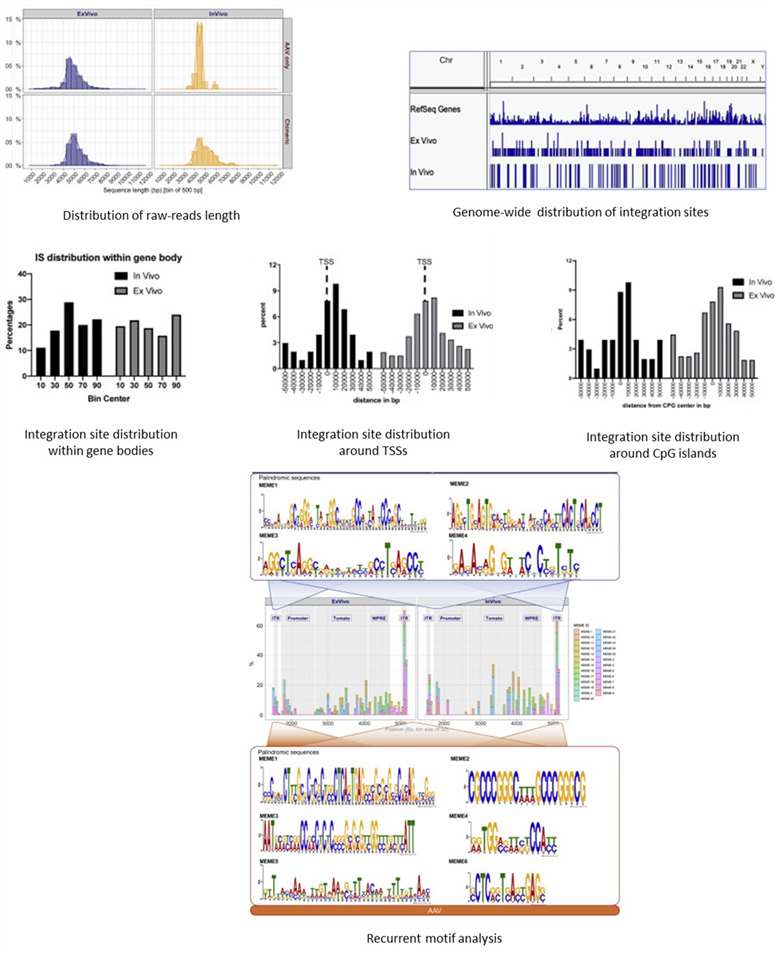
Reference
- Dalwadi D A, Calabria A, Tiyaboonchai A, et al. AAV integration in human hepatocytes. Molecular Therapy, 2021, 29(10): 2898-2909.
1. What are the benefits of AAV integration site analysis?
AAV Integration Site Analysis characterizes integration patterns, assesses safety, refines approaches, and supports regulatory compliance.
2. How does CD Genomics conduct AAV integration site analysis?
The process involves sample collection, DNA extraction, library preparation, sequencing, bioinformatics analysis, and data interpretation.
3. How is the quality of sequencing data ensured?
Quality control measures, such as FastQC, are employed to verify sequencing data quality before analysis.
4. Can integration site analysis predict potential safety concerns?
The analysis identifies integration hotspots and potential oncogenic insertions, helping to assess risks associated with insertional mutagenesis.
Integrated vector genomes may contribute to long-term expression in primate liver after AAV administration
Journal: Nature Biotechnology
Impact factor: 68.164
Published: 06 November 2023
Background
Liver-based AAV approaches initially achieve high transgene expression in nonhuman primates but experience a decline over 90 days, stabilizing at lower levels. The study finds that AAV-mediated expression involves a short-lived phase from episomal genomes and a long-term phase from integrated vectors, with genomic integrations detected in a small fraction of cells.
Materials & Methods
Sample Preparation
- AAV vector production
- Adult cynomolgus
- Rhesus macaques
- Tissue samples
- DNA extraction
- RNA extraction
Method
- IHC for CG
- ISH for vector DNA and transgene RNA
- snRNA-seq
- Next-generation sequencing for AAV integration analysis
- long-read sequencing
- Oxford nanopore sequencing
- Reference genome alignment
- Principal component analysis
- UMAP
- AAV integration site identification
- Integration site annotation
Results
The study examined factors affecting transgene expression in liver AAV systems using rhesus macaques and β-choriogonadotropic glycoprotein (rh-β-CG) as a transgene. It assessed AAV8 and AAVrh10 vectors through liver biopsies at various time points. Findings indicated that rh-β-CG protein levels peaked early and stabilized at lower levels, while vector DNA and RNA decreased over time, with RNA remaining stable in later assessments.
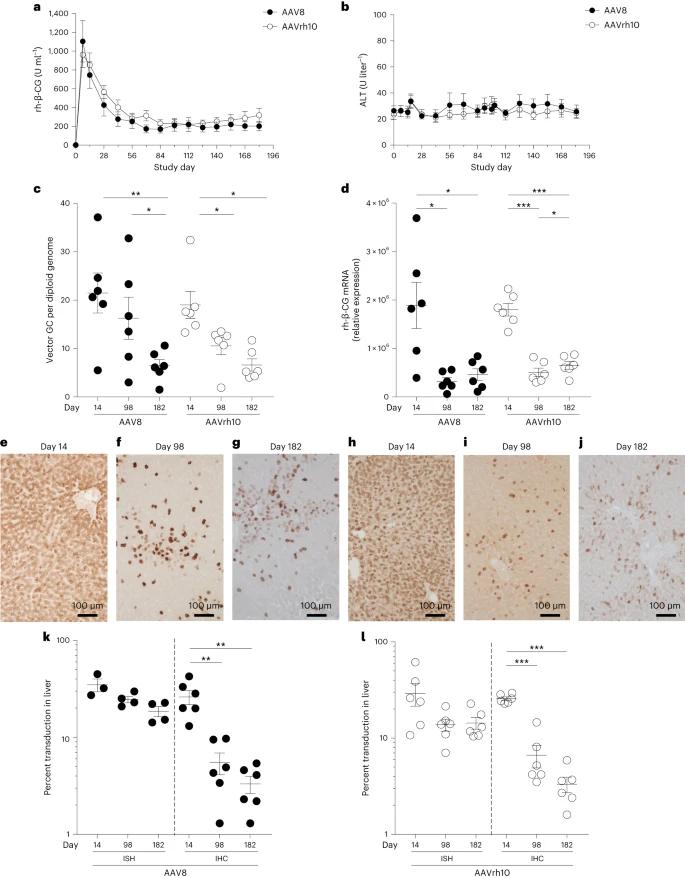 Fig. 1: Initial peak followed by a decline to lower stable levels of self-transgene after i.v. administration of AAV vectors to NHPs.
Fig. 1: Initial peak followed by a decline to lower stable levels of self-transgene after i.v. administration of AAV vectors to NHPs.
The study found that AAV vectors integrate into chromosomal DNA as complex concatemeric structures, with high integration frequencies observed in non-expressing hepatocytes. Using inverted terminal repeat sequencing (ITR-seq), researchers identified integration events in liver DNA, revealing a widespread distribution across the genome, particularly near highly expressed liver genes. Notably, none of the integration sites were located in regions associated with hepatocellular carcinoma mutations. Clonal expansions were analyzed, indicating that animals exposed to certain vectors had more expanded loci. Additionally, long-read sequencing demonstrated a diverse array of AAV sequences in vivo, highlighting rearrangements and truncations that likely occurred post-administration.
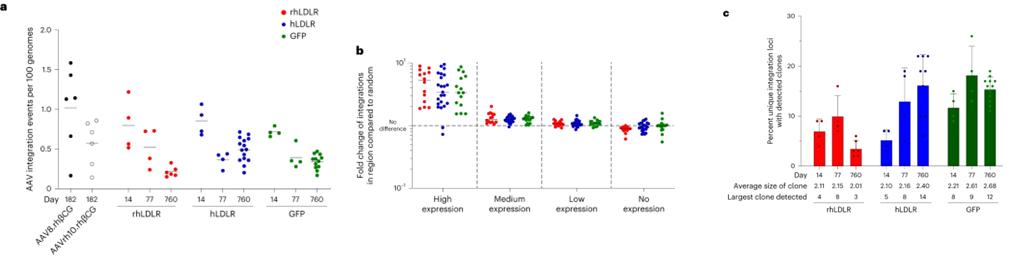 Fig. 2: Localization of integrated vector DNA following i.v. administration of AAV vectors to NHPs.
Fig. 2: Localization of integrated vector DNA following i.v. administration of AAV vectors to NHPs.
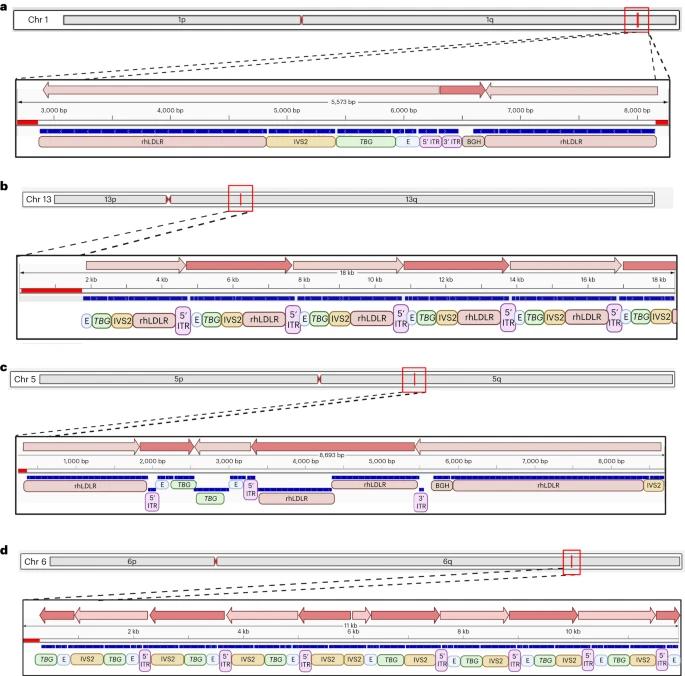 Fig. 3: Structure of integrated vector DNA following i.v. administration of AAV vectors to NHPs.
Fig. 3: Structure of integrated vector DNA following i.v. administration of AAV vectors to NHPs.
Conclusion
The study shows that AAV in primate livers is influenced by immune responses and DNA repair. Persistent nuclear domains of vector DNA exist alongside decreasing transgene expression, indicating potential silencing. Low-frequency integrations suggest early expression depends on episomal forms. Enhancing effectiveness may involve promoting episomal activity and targeting integrations for durability.
Reference
- Greig J A, Martins K M, Breton C, et al. Integrated vector genomes may contribute to long-term expression in primate liver after AAV administration. Nature Biotechnology, 2023: 1-11.
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
