HIV (Human Immunodeficiency Virus) is a retrovirus that belongs to the lentivirus family. It mainly destroys the human immune system by destroying CD4 cells, leading to the occurrence of AIDS. Research on the HIV genome is crucial to understanding the virus's replication mechanism, the infection process, and the development of antiviral treatments. This article aims to comprehensively introduce the genomic structure, composition and biological significance of the HIV virus, and help readers better understand the infection mechanism and treatment strategies of HIV.
Basic Structure of the HIV Genome
The Structure of the HIV Genome
HIV is an RNA virus. Although its genome will eventually be transformed into DNA and integrated into the host cell's chromosomes, its original form is the RNA genome. The genome of the HIV virus is a single-segment, linear, dimeric, single-stranded positive-sense RNA with a length of 9.75 kb and a 5'cap structure and a 3' polyadenylate tail. There is a long terminal repeat (LTR) of approximately 600nt at each end of the genome, which includes the U3, R and U5 regions. In addition, the 5'end also contains a primer binding site (PBS) and the 3' end contains a polypurine sequence (PPT). These structural characteristics are crucial to the virus's replication and infection process.
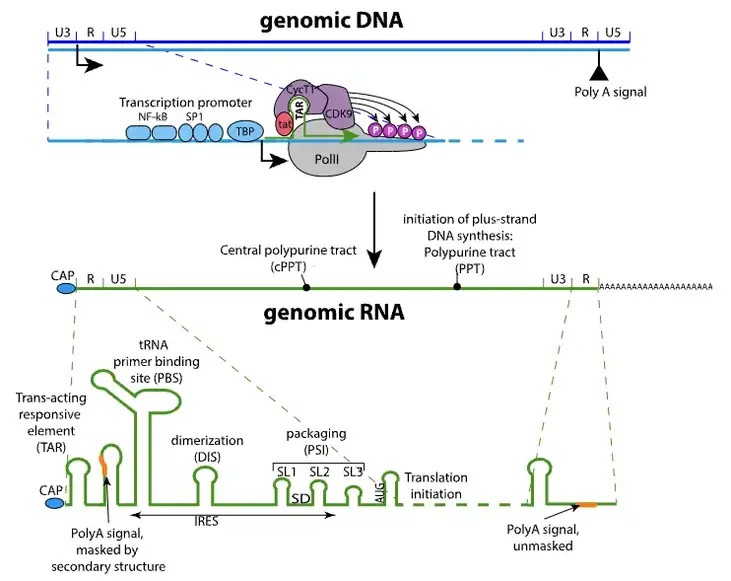 Figure1. HIV Virus Genome and Proteins .(Image Source: https://viralzone.expasy.org/5183)
Figure1. HIV Virus Genome and Proteins .(Image Source: https://viralzone.expasy.org/5183)
Service you may interested in
Resources
Is the HIV Genome Double-Stranded?
The HIV genome is not double-stranded. HIV is a retrovirus whose genome consists of two identical single-stranded RNA molecules wrapped in the core of the virus particle. After infecting a host cell, the virus's single-stranded RNA is transcribed into double-stranded DNA (dsDNA) through the action of reverse transcriptase, a critical step in HIV replication. Therefore, the HIV genome is in the form of single-stranded RNA rather than double-stranded before infecting the host cell.
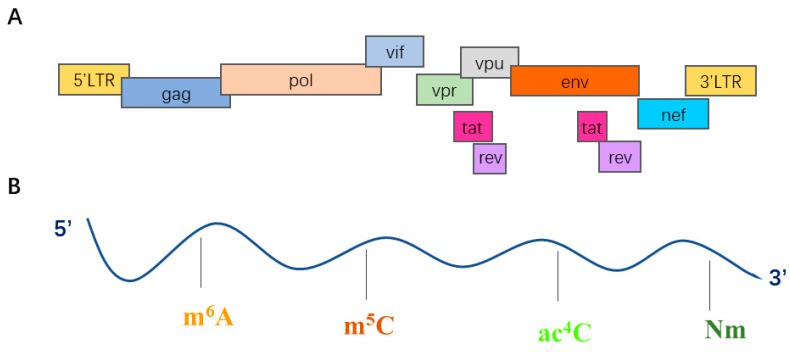 Figure 2 .Structure and organization of the HIV-1 genome.(Wang, S.,et.al,2022)
Figure 2 .Structure and organization of the HIV-1 genome.(Wang, S.,et.al,2022)
Is HIV Genome Positive or Negative?
The genome of HIV is positive-sense RNA. This means that HIV RNA can be directly used as a translation template for synthesizing viral proteins and mRNA. The characteristic of positive-strand RNA is that its sequence is similar to mRNA and can be directly recognized and translated into protein by the host cell's translation mechanism. However, during the life cycle of HIV, positive-strand RNA needs to undergo a process of reverse transcription, which converts it into complementary DNA, then forms double-strand DNA (dsDNA), and ultimately integrates into the host cell's chromosomes.
Detailed Structure of the HIV Genome
Genome Composition
Monopartite, Linear, Dimeric, ssRNA(+)
The HIV genome consists of two single-stranded positive-strand RNAs that are bonded by hydrogen bonds to form a dimer. This structure allows the virus to use the ribosomes of the host cell for translation and synthesis of the proteins needed by the virus.
Long Terminal Repeats (LTRs)
The HIV genome has an LTR region approximately 300-400 bases long at the 5'and 3'ends. The LTR region contains promoters, enhancers and negative regulatory regions, which are critical for the expression of viral genes.
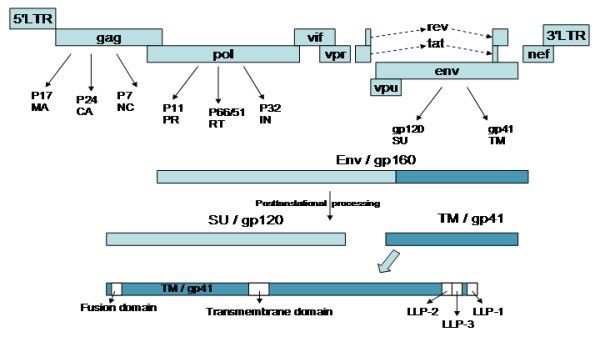 Figure 3 .HIV genome and replication cycle.(Costin J. M, 2007)
Figure 3 .HIV genome and replication cycle.(Costin J. M, 2007)
Primer Binding Site (PBS) and Polypurine Tract (PPT)
PBS is located in the 5'LTR region, which is the starting point for reverse transcriptase to bind to the RNA template to synthesize cDNA;PPT is located in the 3'LTR region, which is involved in the transcription termination and processing of viral RNA.
Genes and Proteins
The HIV genome encodes at least 9 structural proteins and 6 accessory proteins. The functions of these genes and proteins are as follows:
Structural genes
HIV's structural genes include three core genes: gag, pol and env. The gag gene encodes the core structural proteins of the virus, including matrix protein (MA/p17), capsid protein (CA/p24), nucleocapsid protein (NC) and other related proteins, which are responsible for the physical assembly of virus particles. The pol gene encodes key enzymes needed for viral replication: protease (PR) cleaves viral precursor proteins, reverse transcriptase (RT) converts viral RNA into DNA, and integrase (IN) inserts viral DNA into the host genome. The env gene encodes the envelope glycoproteins SU (such as gp120) and TM (such as gp41), which mediate the binding of the virus to host cell receptors and membrane fusion.
Accessory Genes
Helper genes optimize infection efficiency by regulating the interaction between the virus and its host. Rev assists in the nuclear export of unspliced viral RNA and promotes late protein expression;Tat enhances viral transcription by binding to RNA;Nef downregulates host immune molecules (such as MHC-I) to evade immune recognition;Vif antagonizes host APOBEC3 enzymes and protects the viral genome;Vpu degrades host restriction factors and promotes virus release;Vpr assists in viral nuclear import and induces cell cycle arrest, creating conditions for viral replication.
HIV Transcription and Splicing
The transcription process of HIV is one of the key steps in its life cycle and involves multiple stages, including initiation, extension, splicing and polyadenylation.
Initiative
After HIV's genomic RNA is integrated into the genome of a host cell, it is recognized and bound to the long terminal repeat (LTR) promoter of viral RNA by the host cell's RNA polymerase II (RNAPII). This process requires the participation of transcription factors such as NF-κB and SP-1, which help recruit P-TEFb (composed of CDK9 and cyclophilin T) to the bulge-loop structure of RNA polymerase II, thereby activating transcription.
Extension
During the transcriptional extension stage, RNA polymerase II moves along the template chain to produce a complete primary transcript (pre-mRNA). During this process, the Tat protein binds to the RNA polymerase II complex, promoting transcriptional elongation. In addition, the Rev protein also plays a role at this stage, helping transport unspliced mRNA from the nucleus to the cytoplasm.
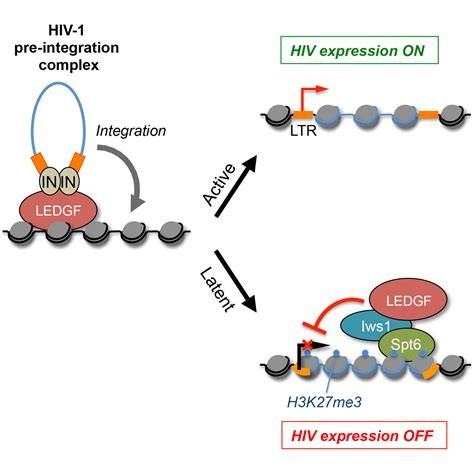 Figure 4 .HIV Extension .(Gérard, Annabelle et al, 2015)
Figure 4 .HIV Extension .(Gérard, Annabelle et al, 2015)
Splicing
HIV's primary transcript (pre-mRNA) goes through a complex splicing process to produce multiple types of transcription products. These transcription products can be divided into the following categories:
Full-length transcript: Contains all sequences encoding structural proteins (such as Gag, Pol, and Gag/Pol) and accessory proteins (such as Rev, Nef)
Single spliced transcript: A sequence that contains accessory proteins such as Vif, Vpr, Vpu, and Env.
Multiple spliced transcripts: sequences that contain only regulatory proteins such as Tat and Rev.
Paused transcript: A transcript that is in a suspended state.
Incomplete transcript: A transcript that has not been spliced or processed.
The splicing process relies on five 5'splicing donor sites (SD1-SD5) and nine 3' splicing acceptor sites (SA1-SA9), which are regulated by conserved donor/acceptor sequences as well as splicing helper sequences such as exon enhancers and intron silencers. The Rev protein is an indispensable factor in the splicing process. It promotes the nuclear export of unspliced mRNA by binding to RRE (Rev Response Element).
Polyadenylation
After splicing is completed, the mRNA undergoes polyadenylation at its 3'end to form a cap structure. This process is accomplished by a series of protein complexes that ensure that mRNA can enter the cytoplasm and be translated into protein.
Nuclear Export
Mature mRNA is transported from the nucleus to the cytoplasm through the nuclear pore complex (NPC). The Rev protein plays a key role in this process, transporting unspliced mRNA from the nucleus to the cytoplasm by binding to RRE.
Genetic Diversity and Evolution
The genetic diversity and evolution of HIV is an important part of its biological characteristics and is mainly driven by the following aspects.
High Mutation Rates
HIV's high genetic diversity mainly stems from the lack of proofreading function of its reverse transcriptase. When reverse transcriptase transcribes viral RNA into DNA, it lacks the 3'-end exonuclease activity and cannot repair errors, resulting in a very high mutation rate. Studies have shown that the mutation rate of HIV ranges from 2×10^-4 to 3.4×10^-4 nucleotides/cycle/second, which means that new mutations are introduced almost every replication (Acevedo-Sáenz, L., et.al,2015).
Nucleotide Composition
HIV's nucleotide composition characteristics (such as rich adenine A and lack of cytosine C) have an important impact on the structure and function of the virus's RNA. This characteristic of nucleotide composition may be related to the stability of the secondary structure of viral RNA. For example, A-rich sequences may be more likely to form stable secondary structures, affecting the efficiency of viral RNA replication and transcription.
Codon Usage and Amino Acid Preferences
HIV's codon use preferences also have an impact on the stability and function of its proteins. Studies have found that HIV-1's codon usage pattern tends to choose A-rich codons, which may be related to the stability of the secondary structure of viral RNA. In addition, the choice of codons may also affect the translation efficiency and expression level of viral proteins.
Conclusion
The HIV genome is like a "double-edged sword"-it is not only the root cause of the virus's ravages, but also the key to humans deciphering its life code. Continuing to deepen the exploration of the biological significance of its genome will not only accelerate the process of ending AIDS, but will also provide a valuable scientific paradigm for dealing with other retroviruses and emerging pathogens.
References:
- Wang, S., Li, H., Lian, Z., & Deng, S. (2022). The Role of RNA Modification in HIV-1 Infection. International journal of molecular sciences, 23(14), 7571. https://doi.org/10.3390/ijms23147571
- Costin J. M. (2007). Cytopathic mechanisms of HIV-1. Virology journal, 4, 100. https://doi.org/10.1186/1743-422X-4-100
- Gérard, Annabelle et al. The Integrase Cofactor LEDGF/p75 Associates with Iws1 and Spt6 for Postintegration Silencing of HIV-1 Gene Expression in Latently Infected Cells.Cell host & microbe, 17(1), 107–117. https://doi.org/10.1016/j.chom.2014.12.002
- Acevedo-Sáenz, L., Ochoa, R., Rugeles, M. T., Olaya-García, P., Velilla-Hernández, P. A., & Diaz, F. J. (2015). Selection pressure in CD8⁺ T-cell epitopes in the pol gene of HIV-1 infected individuals in Colombia. A bioinformatic approach. Viruses, 7(3), 1313–1331. https://doi.org/10.3390/v7031313


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure1. HIV Virus Genome and Proteins .(Image Source: https://viralzone.expasy.org/5183)
Figure1. HIV Virus Genome and Proteins .(Image Source: https://viralzone.expasy.org/5183) Figure 2 .Structure and organization of the HIV-1 genome.(Wang, S.,et.al,2022)
Figure 2 .Structure and organization of the HIV-1 genome.(Wang, S.,et.al,2022) Figure 3 .HIV genome and replication cycle.(Costin J. M, 2007)
Figure 3 .HIV genome and replication cycle.(Costin J. M, 2007) Figure 4 .HIV Extension .(Gérard, Annabelle et al, 2015)
Figure 4 .HIV Extension .(Gérard, Annabelle et al, 2015)