Within epigenetic mechanisms, the methylation of DNA stands as a fundamental regulatory process where methyl groups attach to cytosine bases' 5' position. Such alterations extend beyond simple gene regulation, fundamentally shaping numerous vital functions: the specialization of cells, growth during embryogenesis, genetic imprinting mechanisms, and deactivation processes of X chromosomes. Research has revealed that disrupted methylation patterns contribute substantially to multiple pathological conditions, spanning from malignancies to neural degeneration and heart-related ailments. Understanding these molecular modifications proves essential for grasping disease mechanisms and their progression pathways.
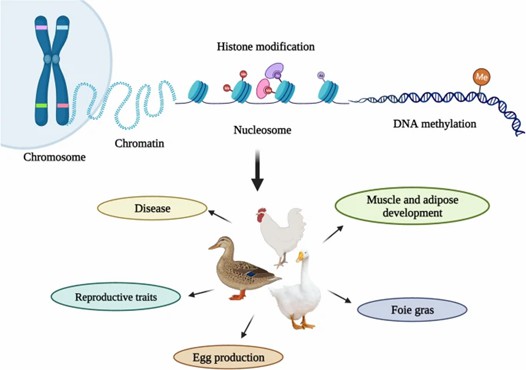 Overview of epigenetic mechanisms in poultry. (Ju, et al., 2023)
Overview of epigenetic mechanisms in poultry. (Ju, et al., 2023)
Modern genomic investigation employs methylation array platforms to examine DNA modifications systematically across entire genomes. Through precise measurement at individual CpG locations, these analytical tools generate detailed epigenetic profiles. The application of array technologies has revolutionized our approach to investigating various medical conditions, yielding crucial insights into distinguishing different cancer types, including mammary and prostatic malignancies. These methodologies have proven invaluable for examining disease patterns across diverse populations, enhancing personalized therapeutic strategies.
Contemporary advances in DNA sequence analysis are transforming methylation research capabilities. Innovative approaches, including sequencing via nanopores and simultaneous analysis methods, create unprecedented possibilities for studying bacterial methylation patterns. Artificial intelligence techniques continue evolving to process extensive methylation datasets effectively. Clinical implementation of these scientific discoveries remains a priority, particularly regarding methylation-based diagnostic indicators for assessing health risks and monitoring treatment outcomes. Scientists are increasingly focusing on understanding how methylation interacts with various epigenetic factors, particularly modifications to histone proteins. Continued scientific investigation into DNA methylation mechanisms promises to advance medical diagnostics while offering novel therapeutic strategies and preventive approaches.
Service you may interested in
Current State of Methylation Arrays
Dominant Array Technologies
Among contemporary epigenetic analysis tools, the Infinium platform developed by Illumina stands at the forefront of DNA methylation detection. Through sophisticated microarray techniques, these systems enable precise quantification of methylation at specific CpG locations. The technology has evolved through multiple iterations, progressing from the initial 27k BeadChip to advanced platforms like the 450k array and EPIC BeadChip, which analyze approximately 485,000 and 850,000 methylation sites, respectively.
The analytical framework employs distinctive chemical approaches - Infinium I and II methodologies - to distinguish methylated from unmethylated genomic regions. While this dual approach yields comprehensive data, it introduces notable analytical complexities.
Alternative Methodological Approaches
Researchers often utilize whole-genome bisulphite sequencing when seeking enhanced precision, despite its resource-intensive nature. The RRBS technique presents an economical alternative, employing targeted enzymatic digestion to analyze specific genomic segments. Commercial solutions from Agilent and Nimblegen offer specialized tools for focused methylation studies in particular genomic regions.
Implementation Domains and Technical Constraints
Research Applications
Methylation arrays have proven invaluable in oncological investigations, revealing distinctive methylation signatures within neoplastic tissues. For example, these platforms have enhanced our understanding of methylation patterns in pulmonary carcinomas. The technology supports broad-ranging epigenetic investigations, including gene regulation mechanisms and chromosomal inactivation studies. Population-based research particularly benefits from EPIC arrays, which examine most RefSeq genes and CpG islands.
Technological Limitations
The integration of dual-probe methodologies creates analytical challenges, particularly regarding probe type variations. Accuracy concerns arise from cross-reactive elements interacting with repetitive DNA sequences or polymorphic regions. Studies highlight potential confusion between probes targeting sex chromosomes and their homologous sequences. Despite their effectiveness in large-scale research, substantial costs restrict widespread implementation. Furthermore, technical variations between samples necessitate sophisticated normalization procedures like Beta-mixture Quantile methods.
While array-based methylation analysis maintains its position as a crucial epigenetic research tool, ongoing refinements must address current technical constraints, data processing challenges, and accessibility concerns to enhance its research applications.
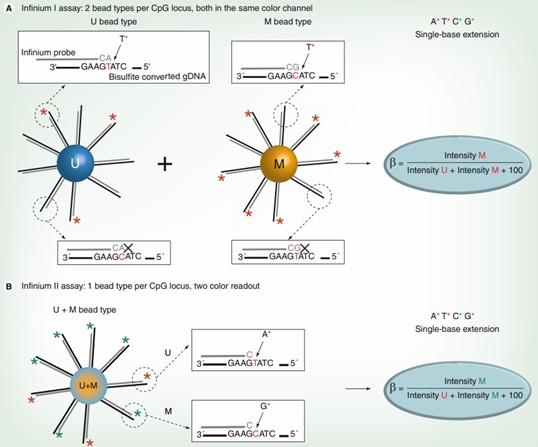 Overview of the Infinium I and Infinium II assays. (Dedeurwaerder, Sarah, et al. 2011)
Overview of the Infinium I and Infinium II assays. (Dedeurwaerder, Sarah, et al. 2011)
Emerging Innovations in Methylation Array Technologies
Recent years have witnessed remarkable advancements in DNA methylation array technologies concerning resolution, coverage, sensitivity, and specificity. The incorporation of NGS technology has not only enhanced data quality but also facilitated deeper epigenetic insights through the integration of novel methodologies such as single-cell analysis and spatial profiling. These innovations serve as powerful tools for disease diagnosis, treatment, and fundamental biological research.
1. Next-Generation Methylation Arrays
Increased Resolution and Coverage
- Next-generation sequencing (NGS) technologies have drastically transformed DNA methylation analysis by facilitating genome-wide profiling at single-base resolution. This advancement enables researchers to discern subtle differences in methylation patterns throughout the genome.
- Among NGS-based methodologies, whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS) are noteworthy for their high-resolution coverage of methylated cytosines across the genome.
Integration with NGS
- NGS is now regarded as the gold standard for DNA methylation analysis due to its capacity for exhaustive, unbiased epigenomic profiling. This technology integrates seamlessly with traditional techniques like bisulfite conversion and restriction enzyme digestion, enhancing both sensitivity and specificity.
- Additionally, targeted NGS panels have been developed for specific genomic regions, allowing researchers to concentrate on particular genes or loci of interest. This focus reduces costs while improving resolution.
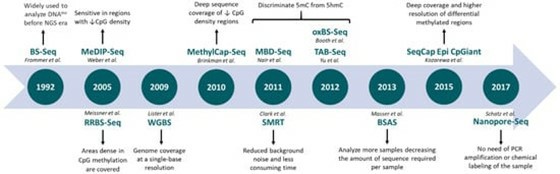 Evolution of next-generation sequencing-based techniques applied to DNA methylation profiling. (Barros-Silva, et al., 2018)
Evolution of next-generation sequencing-based techniques applied to DNA methylation profiling. (Barros-Silva, et al., 2018)
2. Technological Advancements
Improved Sensitivity and Specificity
- Compared to older technologies like microarrays, NGS-based approaches such as RRBS and WGBS have markedly enhanced the sensitivity and specificity of DNA methylation detection. These techniques efficiently and accurately quantify methylation levels at individual cytosines.
- The advent of novel sequencing platforms, including ion torrent sequencing, has further augmented the throughput and precision of methylation analysis.
Enhanced Data Quality and Reproducibility
- Over time, the stability and quality of NGS data have significantly improved, facilitating more reliable dataset comparisons. This is particularly crucial for clinical applications where reproducibility is paramount.
- Established quality control measures, encompassing read alignment, methylation scoring, and feature annotation, have become standard practices in NGS-based methylation studies.
3. New Platforms and Methodologies
Single-Cell Methylation Analysis
- Single-cell sequencing technologies have pioneered the analysis of DNA methylation at the single-cell level, offering insights into cellular heterogeneity and epigenetic regulation within tissues.
- These methodologies are especially valuable in cancer research, assisting in the identification of methylation changes linked to particular cell types or subpopulations.
Spatial Methylation Profiling
- Spatial analysis techniques allow for the mapping of DNA methylation patterns across specific genomic regions or even entire genomes. This approach fosters a deeper understanding of the spatial organization of the epigenome and its functional implications.
- Increasingly, spatial analysis tools are being integrated with NGS data to yield a more comprehensive overview of epigenetic regulation.
Service you may interested in
Want to know more about the details of DNA Methylation Arrays? Check out these articles:
Future Applications of Methylation Arrays
Array-based methylation detection technologies demonstrate remarkable promise across medical practice and scientific investigation. These platforms increasingly influence diagnostic procedures, treatment strategies, and individualized therapeutic approaches. Their impact extends through developmental studies, environmental research, and mechanistic disease investigations. The technology shows particular promise in identifying novel biomarkers and advancing pharmaceutical development.
1. Medical Implementation
Diagnostic and Prognostic Applications
- Specific methylation signatures serve as vital diagnostic and prognostic indicators. Examining methylation patterns within gene regulatory regions enables early cancer detection and outcome prediction. For example, glioma assessment relies heavily on MGMT promoter methylation status. Similarly, altered SOCS3 promoter methylation provides a minimally invasive marker for prostatic malignancies.
- Long-term condition monitoring benefits from methylation-based screening protocols, enabling treatment selection and post-surgical surveillance.
Precision Healthcare Strategies
- Methylation profile analysis creates opportunities for tailored medical interventions. By examining tumor-specific methylation signatures, clinicians can develop patient-specific treatment protocols. These markers additionally help predict therapeutic responsiveness, optimizing clinical outcomes.
2. Scientific Applications
Developmental and Age-Related Studies
- These platforms reveal crucial insights into genetic regulation and epigenetic modifications. Scientists track methylation changes throughout organismal development and aging processes to understand fundamental biological mechanisms.
- Such analyses prove essential for investigating neurodegenerative conditions and autoimmune disorders.
Environmental Impact Assessment
- External factors including nutrition, behavior patterns, and chemical exposure influence health through methylation alterations. Scientists employ methylation analysis to evaluate health impacts of tobacco use, dietary choices, and various environmental elements.
- Exploring connections between external factors and methylation patterns helps reveal chronic disease origins and supports preventive approaches.
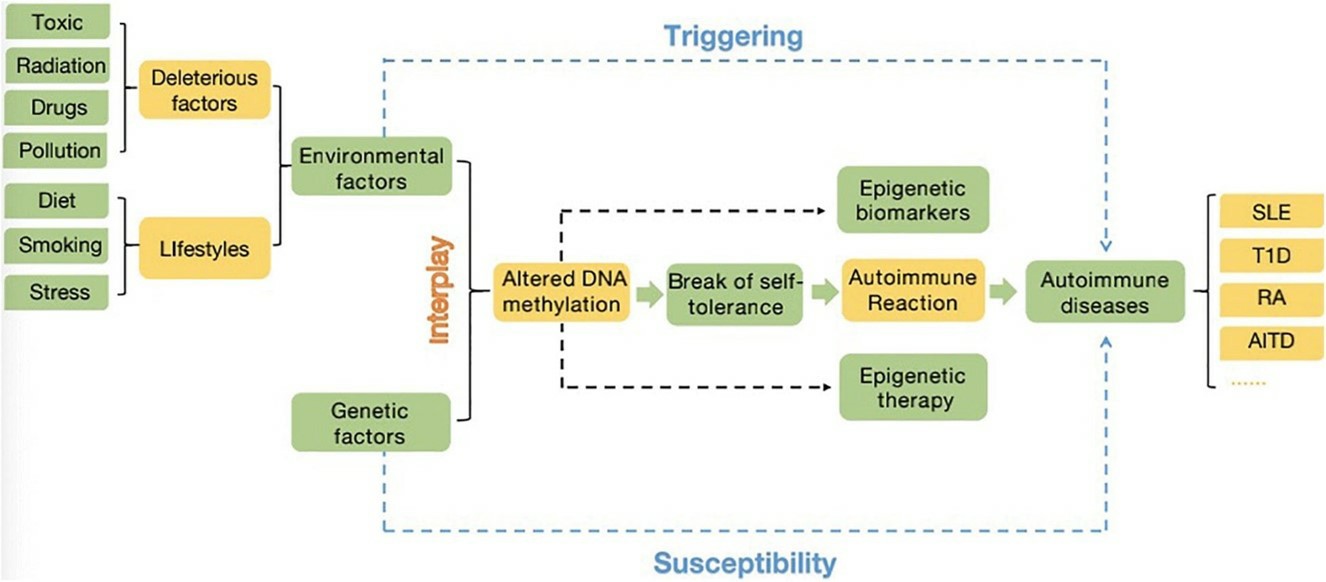 A close interplay between the environment (deleterious factors and lifestyles) and genetic factors (related genes, SNPs, copy number variation…). (Li, Jiaqi, et al., 2021)
A close interplay between the environment (deleterious factors and lifestyles) and genetic factors (related genes, SNPs, copy number variation…). (Li, Jiaqi, et al., 2021)
3. Bridging Research and Clinical Practice
Novel Biomarker Identification
- High-throughput methylation analysis enables discovery of disease-specific indicators. Scientists can identify distinctive methylation patterns associated with particular conditions, supporting early detection and progression monitoring.
- Within oncology, methylation markers help distinguish between cancerous and non-cancerous growths, enhancing screening effectiveness.
Therapeutic Innovation
- Methylation analysis supports development of epigenetically targeted pharmaceuticals. Contemporary techniques combining CRISPR technology with methylation-focused approaches advance precision medicine development.
- Understanding relationships between methylation states and treatment outcomes helps predict drug effectiveness and enhance therapeutic strategies.
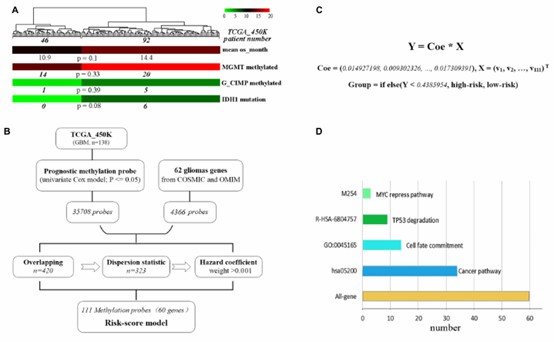 The development of the methylation risk-score signature. (Chen, Qi, et al. 2019)
The development of the methylation risk-score signature. (Chen, Qi, et al. 2019)
These technological advances have substantially improved detection precision, genomic coverage, and measurement accuracy. Integration with contemporary sequencing methods enhances data reliability, while innovative approaches examining individual cells and spatial relationships provide deeper epigenetic understanding. Such developments strengthen both clinical applications and fundamental biological research.
If you want to learn more, you can read the following articles:
Case Studies and Future Directions
Examples of Innovative Methylation Array Applications
Methylation arrays have been extensively adopted across various research domains, notably in cancer research and studies of human diseases. These arrays facilitate the examination of epigenetic modifications throughout the genome with single-nucleotide precision, offering extensive coverage of regions like CpG islands, CHH sites, enhancers, open chromatin, and transcription factor binding sites. Their user-friendly nature and capability for high-throughput analysis make them particularly suitable for handling large sample sets with minimal input requirements. Methylation arrays have also been instrumental in identifying novel biomarkers for diseases such as hepatocellular carcinoma (HCC) and cervical cancer, aiding in the detection of clinically significant methylation alterations.
Potential Impact on Scientific and Medical Advancements
The deployment of methylation arrays holds profound implications for scientific discovery and medical practice alike. In the realm of cancer research, these arrays have facilitated the identification of disease-specific methylomes, contributing to the refinement of cancer subtype classification and enhancing diagnostic precision. Moreover, methylation arrays support the advancement of personalized medicine by allowing for the customization of treatment strategies based on individual epigenetic profiles. Beyond oncology, methylation arrays show promise in fields such as autoimmune diseases, where they help in identifying biomarkers relevant to disease risk assessment and therapeutic monitoring. The synergistic integration of methylation data with other omics technologies further enriches our comprehension of complex biological processes and disease mechanisms.
Ethical and Regulatory Considerations
Despite the considerable benefits offered by methylation arrays, their utilization introduces several ethical and regulatory challenges. A key concern is the potential for bias in methylation data due to variables such as bisulfite conversion efficiency or probe hybridization conditions. To mitigate these issues, it is essential to develop standardized protocols and validate analytical pipelines to ensure reproducibility and robustness across studies. Additionally, the application of methylation arrays in clinical settings requires meticulous attention to patient privacy and informed consent, particularly when handling sensitive biological samples. Regulatory authorities must also establish clear guidelines concerning the use of methylation-based biomarkers in diagnostic and therapeutic applications to safeguard their safety and efficacy.
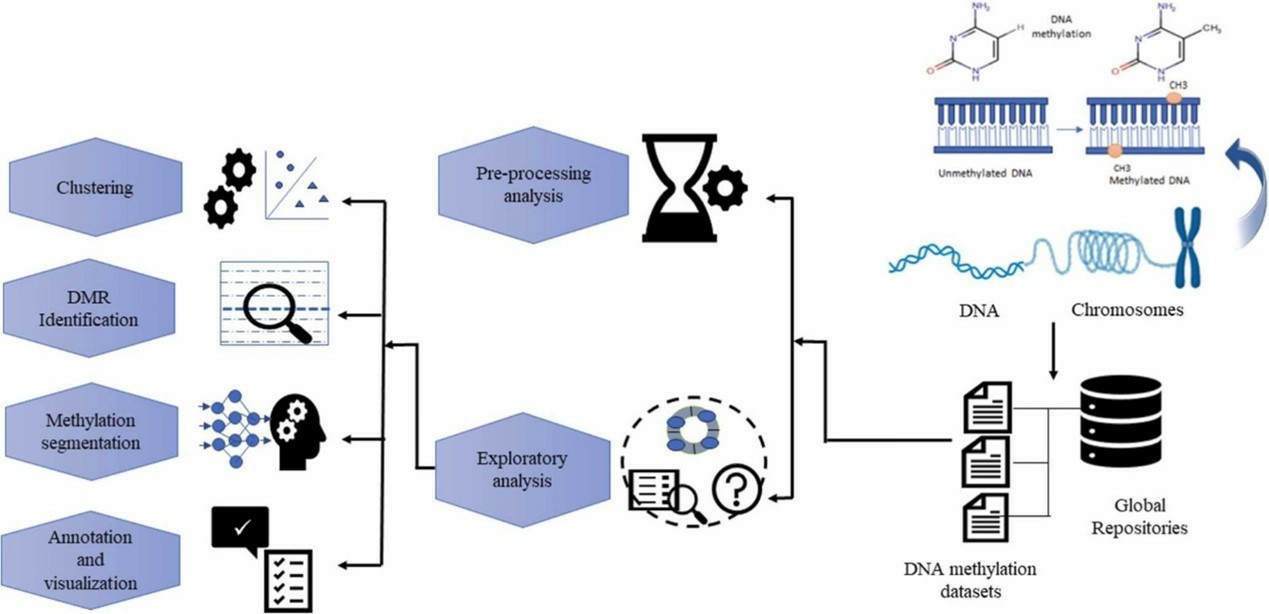 Methods in DNA methylation array dataset analysis: A review. (Sahoo, et al., 2024).
Methods in DNA methylation array dataset analysis: A review. (Sahoo, et al., 2024).
Conclusion
Array-based methylation analysis stands at the forefront of transforming both individualized healthcare and epigenetic research. The convergence of advanced sequencing platforms, including nanopore technologies and comprehensive molecular profiling approaches, has revolutionized our capacity to examine methylation patterns across entire genomes with unprecedented accuracy. Such technological progress promises to reshape our approach to detecting and treating various pathological conditions, with particular significance in oncological applications.
The field's evolution benefits substantially from cross-disciplinary partnerships, unified analytical protocols, and enhanced data accessibility. This collaborative framework strengthens the connection between laboratory discoveries and medical implementation. Through these integrated efforts, researchers gain deeper insights into disease pathogenesis, enabling earlier detection strategies and innovative therapeutic interventions. As these methodologies continue evolving, their impact on advancing personalized medicine and understanding epigenetic regulation grows increasingly significant.
References:
- Ju, X., Wang, Z., Cai, D. et al. DNA methylation in poultry: a review. J Animal Sci Biotechnol 14, 138 (2023). https://doi.org/10.1186/s40104-023-00939-9
- Wu, Jiayun, et al. "Genome-wide DNA methylome and transcriptome analysis of porcine testicular cells infected with transmissible gastroenteritis virus." Frontiers in Veterinary Science 8 (2022): 779323. https://doi.org/10.3389/fvets.2021.779323
- Yassi, Maryam, Aniruddha Chatterjee, and Matthew Parry. "Application of deep learning in cancer epigenetics through DNA methylation analysis." Briefings in bioinformatics 24.6 (2023): bbad411. https://doi.org/10.1093/bib/bbad411
- Maksimovic, Jovana, Belinda Phipson, and Alicia Oshlack. "A cross-package Bioconductor workflow for analysing methylation array data." F1000Research 5 (2017): 1281. doi: 10.12688/f1000research.8839.3
- Dedeurwaerder, Sarah, et al. "Evaluation of the Infinium Methylation 450K technology." Epigenomics 3.6 (2011): 771-784. https://doi.org/10.2217/epi.11.105
- Peters, T.J., Buckley, M.J., Statham, A.L. et al. De novo identification of differentially methylated regions in the human genome. Epigenetics & Chromatin 8, 6 (2015). https://doi.org/10.1186/1756-8935-8-6
- Barros-Silva, Daniela, et al. "Profiling DNA methylation based on next-generation sequencing approaches: new insights and clinical applications." Genes 9.9 (2018): 429. https://doi.org/10.3390/genes9090429
- Li, Jiaqi, et al. "Insights into the role of DNA methylation in immune cell development and autoimmune disease." Frontiers in Cell and Developmental Biology 9 (2021): 757318. https://doi.org/10.3389/fcell.2021.757318
- Chen, Qi, et al. "Hypomethylation of 111 probes predicts poor prognosis for glioblastoma." Frontiers in Neuroscience 13 (2019): 1137. https://doi.org/10.3389/fnins.2019.01137
- Shih, YL., Kuo, CC., Yan, MD. et al. Quantitative methylation analysis reveals distinct association between PAX6 methylation and clinical characteristics with different viral infections in hepatocellular carcinoma. Clin Epigenet 8, 41 (2016). https://doi.org/10.1186/s13148-016-0208-3
- Sahoo, Karishma, and Vino Sundararajan. "Methods in DNA methylation array dataset analysis: A review." Computational and Structural Biotechnology Journal (2024). https://doi.org/10.1016/j.csbj.2024.05.015


 Sample Submission Guidelines
Sample Submission Guidelines Overview of epigenetic mechanisms in poultry. (Ju, et al., 2023)
Overview of epigenetic mechanisms in poultry. (Ju, et al., 2023) Overview of the Infinium I and Infinium II assays. (Dedeurwaerder, Sarah, et al. 2011)
Overview of the Infinium I and Infinium II assays. (Dedeurwaerder, Sarah, et al. 2011) Evolution of next-generation sequencing-based techniques applied to DNA methylation profiling. (Barros-Silva, et al., 2018)
Evolution of next-generation sequencing-based techniques applied to DNA methylation profiling. (Barros-Silva, et al., 2018) A close interplay between the environment (deleterious factors and lifestyles) and genetic factors (related genes, SNPs, copy number variation…). (Li, Jiaqi, et al., 2021)
A close interplay between the environment (deleterious factors and lifestyles) and genetic factors (related genes, SNPs, copy number variation…). (Li, Jiaqi, et al., 2021) The development of the methylation risk-score signature. (Chen, Qi, et al. 2019)
The development of the methylation risk-score signature. (Chen, Qi, et al. 2019) Methods in DNA methylation array dataset analysis: A review. (Sahoo, et al., 2024).
Methods in DNA methylation array dataset analysis: A review. (Sahoo, et al., 2024).