Single cell sequencing technology can reveal cell heterogeneity and provide unprecedented perspectives on cell functions, interactions, and disease mechanisms. 10x Genomics is a leader in single cell sequencing. Its Chromium platform combines microfluidic technology and droplet packaging technology to achieve high-throughput sequencing of single cells. This article aims to introduce the basic principles, experimental procedures and applications in different fields of 10x Genomics single cell sequencing technology.
Principles of 10x Genomics Single-Cell Technology
10x Genomics' single cell sequencing technology is based on microfluidic technology and enables high-throughput, high-precision single cell analysis through the "Gel Beads-in-Emulsion (GEM)" structure. The following is a detailed analysis of the technical principles:
What is single cell sequencing
Single cell sequencing technology aims to sequence and analyze the genome, transcriptome or epigenome of a single cell to reveal cell heterogeneity and complexity. 10x Genomics 'Chromium platform is currently one of the most widely used single cell sequencing platforms. Its core lies in the use of microfluidic technology to encapsulate single cells and reaction reagents in nanoscale droplets, thereby achieving high-throughput single cell capture and analysis.
Service you may interested in
Core technologies of the Chromium platform
The core technologies of the 10x Genomics Chromium platform include the following aspects:
Microfluidic technology: The Chromium platform uses a water-in-oil (O/W)-based microfluidic system that mixes single cells, primers, enzymes and reverse transcription reagents to form GEMs. Each GEM contains a single cell and its transcriptome or genomic information.
GEM structure: GEM is a drop-like microreactor consisting of a bead (Gel Bead) and a water-in-oil emulsion. The bead surface is coated with a coded barcode (10x barcode), molecular barcode and RT primers for subsequent barcode decoding and transcript quantification.
10x barcode system: The beads within each GEM carry a molecular barcode that is used to distinguish sequencing data from different cells. These barcodes bind to unique barcode to ensure the uniqueness of each transcript.
High-throughput capture: The Chromium platform can process thousands to tens of thousands of single cells simultaneously, and each chip can capture up to 80,000 cells, significantly improving the efficiency of single cell sequencing.
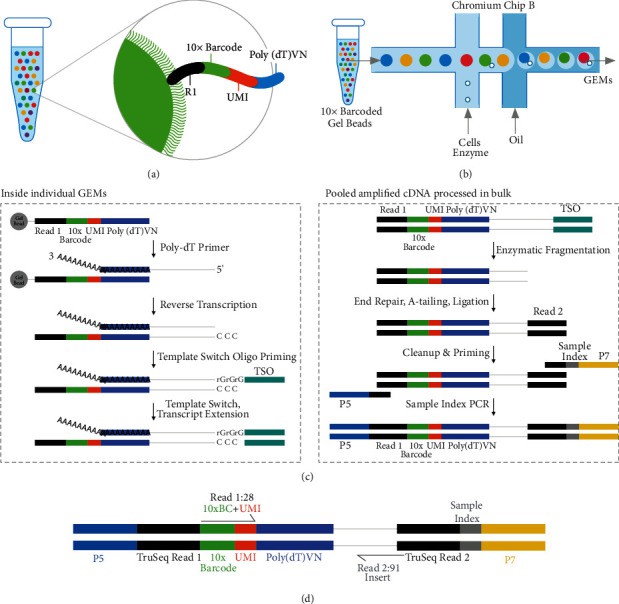 Figure1 .10X genomics working principle process.(Li, Z.,et.al,2022)
Figure1 .10X genomics working principle process.(Li, Z.,et.al,2022)
The workflow of the GEM structure
Droplet generation: After mixing single cell suspension with beads and reverse transcription reagent, GEMs are generated through a microfluidic chip. Each GEM encapsulates a single cell and its transcriptome or genomic information.
Cell lysis and transcription: Within GEM, single cells are lysed, mRNA is reverse transcribed into cDNA, and transcripts are labeled by molecular barcode.
Barcode labeling: The barcodes on the beads are combined with cDNA for subsequent barcode decoding and data classification.
Sequencing library construction: The cDNA in GEM is amplified and sequenced library construction to generate DNA fragments that can be used for sequencing.
Common types of single cell sequencing
The 10x Genomics Chromium platform supports multiple single-cell sequencing types, including:
Single cell RNA sequencing (scRNA-seq): Used to analyze the transcriptome of individual cells, revealing changes in cell type and state.
Single cell ATAC sequencing (scATAC-seq): Used to study the chromatin accessibility of individual cells and analyze gene regulation mechanisms.
Single-cell multimomic sequencing (scMultiome): combines transcriptome and epigenomic information to comprehensively analyze the functional status of individual cells.
Workflow of 10x Genomics Single-Cell Sequencing
The 10x Genomics single cell sequencing workflow is a highly refined and systematic process that mainly includes the following key steps: sample preparation, single cell capture and barcode labeling, library construction, high-throughput sequencing, and bioinformatics analysis. Each link is interconnected to ensure efficient operation of the entire process from sample preparation to data analysis, thereby providing high-resolution single-cell transcriptome data.
Sample preparation
Cell isolation and counting: First, a single cell suspension needs to be separated from tissue or cultured cells. Devices such as the Countess II Automated Cell Counter are usually used to count cells to ensure that cell concentrations are between 700-1200 cells/μL to ensure optimal recovery.
Cell activity assessment: Assess cell activity by staining or flow cytometry and other methods to ensure that the cell survival rate is higher than 80%.
Single cell capture and barcode labeling
Single cell capture: A single cell suspension is mixed with barcoded beads (Gel Beads), and the single cells are dispensed into water-in-oil droplets containing the beads through a microfluidic chip to form GEMs.
Barcode allocation mechanism: The beads in each droplet contain a unique 10x Barcode that is used to subsequently distinguish between different single cell sources.
Library construction
cDNA synthesis and amplification: In droplets, the beads transcribe mRNA into cDNA through reverse transcriptase and use PCR amplification to generate a cDNA library.
Library purification and quality control: Purify and quality test the amplified cDNA library to ensure the integrity of the library.
High-throughput sequencing
Selection of sequencing platform: Sequencing using Illumina platform, usually using equipment such as HiSeq X Ten or NextSeq 550 to generate 150 bp PE reads.
Bioinformatics analysis
10x Cell Ranger analysis pipeline: Cell Ranger software is used to demultiplex, barcode allocation, molecular barcode counting and other processes to generate a gene expression matrix.
Dimension reduction and cluster analysis: Use tools such as Seurat or Scanpy to perform dimension reduction (such as t-SNE or UMAP) and cluster analysis on gene expression data to identify different cell types.
Cell annotation and differential gene expression analysis: Study functional differences among different cell types by annotating known genes or reference genomes, combined with differential expression analysis (such as DESeq2 or edgeR).
Applications of 10x Genomics Single-Cell Technology
10x Genomics single-cell technology has a wide range of applications in basic research, disease research, immunology, and drug development. It is a powerful tool for analyzing cellular heterogeneity, tissue development, and cell fate determination.
Basic Research
Cellular Heterogeneity Analysis: Single-cell sequencing can identify different cell subtypes and analyze the heterogeneity and clustering of cells, including neuron cells and stem cells.
Tissue Development and Cell Fate Determination: It helps in understanding the development of mammalian organs and tissues and the construction of cell lineages based on the characteristics of different cell subtypes(Gupta, R.,etal,2020).
Disease Research
Single-Cell Profiling in Cancer: Single-cell sequencing is used to recognize different cell subtypes within tumors, study tumor cell heterogeneity, and identify new pathogenic pathways and mechanisms. It can characterize tumor cells, stromal cells, and infiltrated immune cells to evaluate tumor progression and responses to environmental changes.
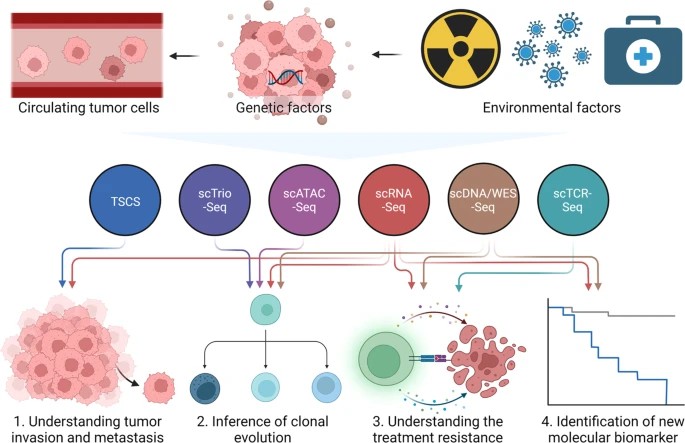 Figure2 .Application of single-cell omics in research of tumor cells.(Jia, Q ,et.al,2022)
Figure2 .Application of single-cell omics in research of tumor cells.(Jia, Q ,et.al,2022)
Neurological Disorders: It enables the analysis of molecular regulation and differentiation mechanisms of neuron cells, aiding in understanding the mechanisms of neural diseases. For brain development research, it helps obtain a gene expression atlas of human brain cells and analyze cell heterogeneity.
Disease Typing: It assists in discovering abnormal cell types to aid in disease typing.
Immunology Research
Immune Cell Lineage Characterization: 10x single-cell technology is used to characterize immune cell lineages by identifying and labeling different subtypes of immune cells and analyzing their genetic diversity. For example, in single-cell RNA sequencing (scRNA-seq) experiments, cell type doublets, which express markers from multiple lineages, can be identified(Majzner, R.,et.al, 2022).These may include T cells (CD2, CD3E, CD3D, CD3G, CD247, CD7, GD2 CAR), B cells (CD19, CD22, PAX5), microglia/myeloid cells (CD14, CD68, CD163, CSF1R, AIF1), or astrocytes (GFAP).
Applications in Immunotherapy: It is useful in tumor immunology and immunotherapy and can help clarify the cellular and molecular mechanisms of immune checkpoint blockade (ICB)-driven tumor control.
Conclusion
The innovative cellular analysis platform developed by 10x Genomics represents a breakthrough in understanding biological complexity at the individual cell level. Through advanced microfluidic systems and precision droplet formation, their Chromium technology facilitates comprehensive examination of vast numbers of discrete cells simultaneously. This methodological approach has transformed researchers' capacity to probe cellular diversity and decode intricate biological mechanisms.
The platform's applications span numerous scientific domains, from fundamental biological investigations to therapeutic development. Researchers studying cancer biology, nervous system disorders, and immune responses have leveraged this technology to generate unprecedented insights. The methodology's particular strength lies in its capacity to analyze thousands of cells while maintaining exceptional resolution at the single-cell level.
What distinguishes this approach is its versatility combined with robust analytical capabilities. By enabling detailed investigation of cellular processes, molecular interactions, and disease pathways, the platform has established new standards in genomic research. This technological advancement continues to accelerate scientific discovery across multiple disciplines, contributing significantly to both basic research and clinical applications.
References:
- Li, Z., Yin, L., Li, Y., Cao, Y., & Zeng, H. (2022). Single-Cell RNA-Sequencing Reveals the Cellular and Genetic Heterogeneity of Skin Scar to Verify the Therapeutic Effects and Mechanism of Action of Dispel-Scar Ointment in Hypertrophic Scar Inhibition. Evidence-based complementary and alternative medicine : eCAM, 2022, 7331164. https://doi.org/10.1155/2022/7331164
- Gupta, R. K., & Kuznicki, J. (2020). Biological and Medical Importance of Cellular Heterogeneity Deciphered by Single-Cell RNA Sequencing. Cells, 9(8), 1751. https://doi.org/10.3390/cells9081751
- Jia, Q., Chu, H., Jin, Z. et al. High-throughput single-сell sequencing in cancer research. Sig Transduct Target Ther 7, 145 (2022). https://doi.org/10.1038/s41392-022-00990-4
- Majzner, R. G., Ramakrishna, S., Yeom, K. W.,et.al. (2022). GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature, 603(7903), 934–941. https://doi.org/10.1038/s41586-022-04489-4


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure1 .10X genomics working principle process.(Li, Z.,et.al,2022)
Figure1 .10X genomics working principle process.(Li, Z.,et.al,2022) Figure2 .Application of single-cell omics in research of tumor cells.(Jia, Q ,et.al,2022)
Figure2 .Application of single-cell omics in research of tumor cells.(Jia, Q ,et.al,2022)