Single-cell isoform sequencing is transforming the field of transcriptomics by delivering unparalleled insights into gene expression and regulation at the single-cell resolution. This sophisticated approach is indispensable for elucidating intricate biological processes and disease mechanisms that are frequently obscured by traditional bulk RNA sequencing.
CD Genomics is leading this innovative frontier by providing state-of-the-art single-cell isoform sequencing services. Through the utilization of advanced technologies such as PacBio MAS-Seq, CD Genomics achieves high-resolution, full-length isoform data, thereby advancing our understanding of cellular heterogeneity and molecular complexity.
What is Single-Cell Isoform Sequencing?
Single-cell isoform sequencing (scIso-Seq) stands as a robust technique for dissecting the transcriptome of individual cells, thereby unveiling the complex nuances of gene expression and splicing variations that underpin cellular diversity and function. In contrast to conventional RNA sequencing, which aggregates gene expression profiles from a population of cells, scIso-Seq preserves the heterogeneity among individual cells. This method facilitates the identification of unique isoforms and splice variants potentially pivotal in health and disease, offering deeper insights into the molecular underpinnings of biological processes.
Importance of Single-Cell Isoform Sequencing
The transcriptome's complexity, defined by various splicing events and isoforms, significantly influences cellular behavior and function. Traditional short-read sequencing methods often fall short in resolving these complexities due to their inability to capture full-length transcripts. In contrast, scIso-Seq provides a comprehensive view of the entire transcript, from the 5' transcription start site to the 3' poly-A tail, uncovering crucial information about exon usage, splice junctions, and untranslated regions (UTRs). This level of detail is essential for accurate gene annotation, understanding regulatory mechanisms, and identifying novel therapeutic targets.
PacBio MAS-Seq for Single-Cell Isoform Sequencing
PacBio's Multiplexed Arrays Sequencing (MAS-Seq) represents a significant advancement in single-cell isoform sequencing technology. By combining full-length cDNA synthesis with long-read sequencing capabilities, MAS-Seq addresses the limitations of traditional short-read methods and enhances the resolution and throughput of single-cell transcriptomics.
Principle of MAS-Seq
MAS-Seq involves the concatenation of full-length cDNA molecules into larger fragments, which are then sequenced using PacBio's high-fidelity (HiFi) long-read technology. The process begins with the generation of single-cell cDNA, followed by the removal of template-switching oligos (TSOs) and the concatenation of cDNA molecules into ordered arrays. These arrays are ligated with MAS adapters and sequenced on PacBio platforms like Sequel II/IIe and Revio systems.
The resulting HiFi reads are bioinformatically segmented to retrieve the original cDNA sequences, ensuring high accuracy and throughput. This method eliminates the need for fragmentation and short-read assembly, providing a direct and comprehensive view of the transcriptome at the single-cell level.
Advantages of MAS-Seq
- Full-Length Isoform Information: MAS-Seq captures complete isoforms, including all exonic regions, splice junctions, and UTRs, providing a detailed map of the transcriptome.
- High Throughput: The concatenation strategy significantly increases throughput, allowing the sequencing of multiple cDNA molecules in a single run. This efficiency is particularly beneficial for large-scale studies and complex tissues.
- Accurate Cell Barcode: MAS-Seq integrates single-cell barcoding and unique tag, ensuring precise quantification and identification of individual transcripts.
- Cost-Effectiveness: By maximizing the use of sequencing capacity, MAS-Seq reduces the cost per cell and per isoform, making it a viable option for extensive single-cell studies.
- Elimination of Orthogonal Sequencing: MAS-Seq provides comprehensive isoform data independently, streamlining the workflow and reducing complexity.
- Enhanced Isoform Detection: MAS-Seq's ability to capture full-length isoforms provides a comprehensive view of alternative splicing events, fusion genes, and other transcriptomic variations often missed by short-read sequencing.
- High-Resolution Data: The long-read technology used in MAS-Seq offers high resolution and accuracy, enabling the identification of subtle transcriptomic changes and rare isoforms critical for understanding cellular functions and disease mechanisms.
- Streamlined Workflow: MAS-Seq simplifies the sequencing workflow, reducing the time, cost, and complexity of single-cell isoform sequencing projects.
- Compatibility and Flexibility: The MAS-Seq kit is compatible with cDNA generated from the 10x Chromium system, making it adaptable to various single-cell sequencing applications. Its flexibility allows researchers to study a wide range of biological samples and experimental conditions.
- Robust Bioinformatics Support: PacBio provides comprehensive bioinformatics tools for processing MAS-Seq data, including read segmentation, unique tag correction, and isoform classification. These tools ensure accurate and efficient data analysis, facilitating downstream applications and insights.
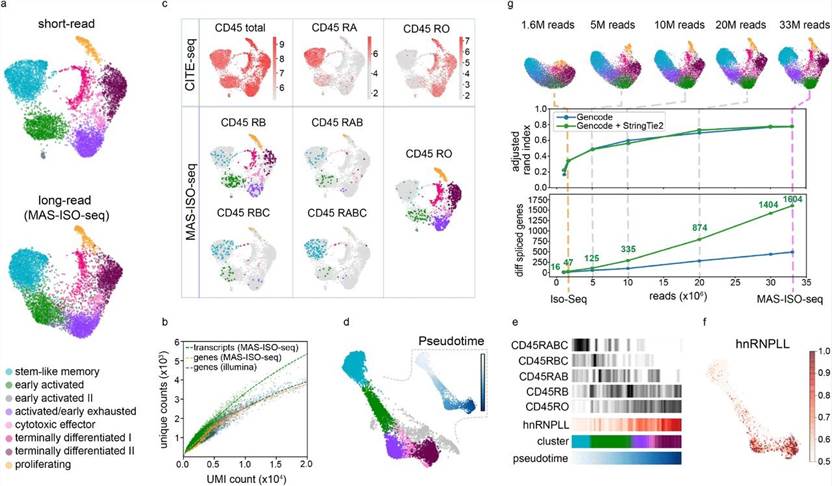 Figure 1. Single-cell isoform-resolved sequencing of single cells with MAS-ISO-seq.
Figure 1. Single-cell isoform-resolved sequencing of single cells with MAS-ISO-seq.
Applications and Impact of MAS-Seq
Single-cell isoform sequencing utilizing MAS-Seq carries significant implications across several domains of biological research and medicine. In the context of cancer research, this technology facilitates the identification of isoform-specific mutations and splicing events that underlie tumorigenesis and therapy resistance. Within neurobiology, it permits a detailed examination of neuronal diversity and the detection of isoform variations linked to neurological disorders. Furthermore, MAS-Seq proves invaluable in plant genomics, aiding in the elucidation of complex transcriptomes in species with extensive and diverse genomes.
Our Single-Cell Isoform Sequencing Service by MAS-Seq
At CD Genomics, we harness the advanced MAS-Seq technology to deliver unparalleled resolution in transcriptomic analyses. Our service captures full-length isoforms, providing a comprehensive understanding of alternative splicing, gene fusion events, and other transcriptomic variations often obscured by conventional sequencing methods.
In addition to our sophisticated isoform sequencing capabilities, we are committed to offering high-quality single-cell RNA sequencing, single-cell DNA sequencing, single-cell DNA methylation sequencing, and 10X Genomics single-cell sequencing. This multipronged platform enables us to cater to a broad spectrum of research needs, furnishing detailed insights into gene expression, genetic variations, and epigenetic modifications at the single-cell level.
MAS-Seq Data Analysis Workflow and Content
| Step | Description | Details |
|---|---|---|
| Data Preprocessing | Convert MAS-Seq data into a usable analysis format and add relevant metadata. | - Quantified and processed RNA capture and antibody capture libraries. - Addressed conservative cell identification to retain more viable droplets. - Removed non-immune cells and doublets. - Log-transformed antibody capture counts for further analysis. |
| Data Integration | Integrate long-read and short-read data to ensure consistency and high-quality joint analysis. | - Converted isoform-level count matrices for analysis. - Mapped novel transcripts and genes to known annotations. - Marked ambiguous transcripts and cross-referenced receptor genes. - Harmonized datasets by retaining mutual cell barcodes, achieving high identification fidelity. |
| Normalization, Clustering, and Embedding | Normalize the data, identify highly variable features, and perform clustering analysis. | - Applied variance-stabilizing transformation to counts. - Selected top genes/isoforms as highly variable features. - Reduced feature set using principal component analysis. - Constructed nearest neighbor graph for embedding and clustering. - Conducted differential gene expression analysis. |
| Differential Expression Analysis | Identify differential expression genes between different cell types and annotate cell populations. | - Performed differential expression analysis to identify expression differences between different cell types. |
| Evolutionary Trajectory Analysis | Use diffusion pseudotime (DPT) analysis to map cell developmental paths. | - Conducted diffusion pseudotime analysis to determine cell state transitions. - Utilized transformed counts for cleaner graphical representation. |
| CD45 Isoform Annotation Refinement | Refine GENCODE annotations to improve specificity of isoform assignments. | - Refined incomplete gene annotations - Improved specificity for isoform assignments using enhanced methods. - Employed decision-based assignment for accurate isoform detection. |
| Identification of Differentially Spliced Genes | Perform global and cluster-resolved differential splicing statistical tests to identify differential isoform usage across cell populations. | - Conducted global differential splicing tests and cluster-resolved differential splicing tests to identify differential isoform usage in different cell clusters. - Adjusted p-values for false discovery rate (FDR) to ensure statistical robustness. |
Workflow of Our Service
Sample Preparation: Single-cell suspensions are prepared and processed using the 10x Chromium Next GEM Single Cell 3' kit to generate cDNA.
cDNA Synthesis and Cleanup: Full-length cDNA molecules are synthesized and cleaned up to remove template-switching oligos (TSOs).
Concatenation and Adapter Ligation: cDNA molecules are concatenated into ordered arrays, and MAS adapters are ligated to facilitate sequencing.
Long-Read Sequencing: The prepared cDNA libraries are sequenced on PacBio platforms, such as Sequel II/IIe or Revio systems, generating high-fidelity long reads.
Data Analysis: The HiFi reads are bioinformatically segmented to retrieve the original cDNA sequences, followed by detailed analysis of isoform content, alternative splicing, and gene fusions.
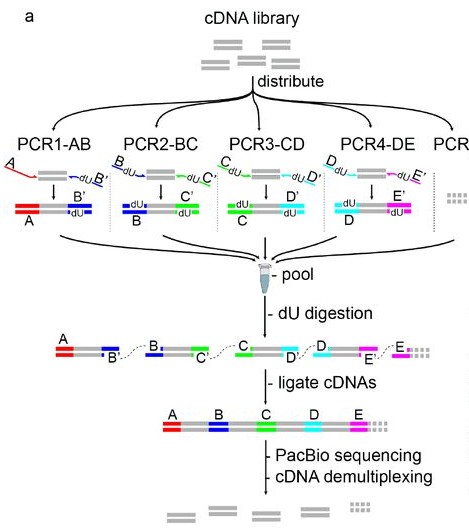 Figure 2. MAS-ISO-seq workflow
Figure 2. MAS-ISO-seq workflow
Sample Requirement
Cell Viability: High cell viability (>90%) is crucial for generating quality single-cell cDNA libraries.
Cell Number: A minimum of 1,000 cells is recommended to achieve comprehensive coverage and accurate isoform quantification.
Sample Purity: Samples should be free from contaminants and debris to prevent interference with the sequencing process.
RNA Integrity: High RNA integrity (RIN > 7) is essential for generating full-length cDNA molecules.
How do you ensure data accuracy and quality?
We utilize PacBio's high-fidelity sequencing technology and robust bioinformatics tools to ensure high accuracy and quality in our data output. Additionally, our expert team performs rigorous quality control checks throughout the workflow.
Can you analyze rare and low-abundance isoforms?
Yes, MAS-Seq is highly sensitive and capable of detecting rare and low-abundance isoforms, providing a comprehensive view of the transcriptome.
What types of samples can you process?
We can process a wide range of single-cell samples, including those from tissues, cell lines, and primary cells. Please contact us for specific sample requirements and recommendations.
What is the minimum number of cells required for single-cell isoform sequencing?
For optimal results, we recommend a minimum of 1,000 cells. This ensures comprehensive coverage and accurate isoform quantification. However, we can work with smaller cell numbers if necessary, though it may impact the depth of analysis.
What type of sequencing platforms do you use for single-cell isoform sequencing?
We use PacBio Sequel II/IIe or Revio systems for single-cell isoform sequencing. These platforms provide high-fidelity long reads, ensuring accurate and detailed isoform analysis.
How do you handle data privacy and security?
At CD Genomics, we prioritize data privacy and security. All data is stored on secure servers with restricted access, and we adhere to strict data protection protocols to ensure the confidentiality of our clients' information.
Can you provide assistance with data interpretation?
Yes, our expert bioinformatics team is available to assist with data interpretation. We offer comprehensive support to help you understand the results, including detailed reports and consultation services.
What sample types are compatible with your single-cell isoform sequencing service?
Our service is compatible with various single-cell samples, including fresh and frozen tissues, cell lines, and primary cells. If you have specific sample types, please contact us for detailed recommendations.
What kind of bioinformatics analysis do you provide?
Our bioinformatics analysis includes unique tag correction, isoform classification, alternative splicing analysis, gene fusion detection, and differential expression analysis. We provide comprehensive reports with visualizations to facilitate data interpretation.
How should samples be prepared and shipped?
Samples should be prepared as single-cell suspensions with high viability and purity. For shipping, we recommend using dry ice and appropriate packaging to maintain sample integrity during transit. Detailed sample preparation and shipping guidelines will be provided upon request.
Can your service identify novel isoforms?
Yes, our single-cell isoform sequencing service is designed to identify novel isoforms. The high-resolution data generated by MAS-Seq allows for the discovery of previously uncharacterized isoforms, enhancing our understanding of the transcriptome.
Is your service suitable for clinical research?
Our service is intended solely for research purposes and is not suitable for clinical diagnosis or testing. However, it is highly valuable for research applications, including cancer research, rare disease studies, and personalized medicine.
What is the cost of your single-cell isoform sequencing service?
The cost of our service varies depending on the project scope, sample number, and specific requirements. Please contact us for a detailed quote tailored to your needs.
Do you offer custom solutions for specific research needs?
Yes, we offer custom solutions to meet specific research needs. Our team will work with you to design a tailored approach that addresses your unique research objectives and challenges.
How can I get started with CD Genomics' single-cell isoform sequencing service?
To get started, simply contact us with your project details and requirements. Our team will provide a consultation to discuss your needs and outline the next steps for sample submission and project initiation.
Reference
- Al'Khafaji, A.M., Smith, J.T., Garimella, K.V. et al. High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat Biotechnol 42, 582–586 (2024).


 Sample Submission Guidelines
Sample Submission Guidelines
