Functions of the Soil Microbiome
A central tenet of Soil microbiomes research is the characterization of the co-evolutionary mechanisms within microbial communities in specific soils, and elucidation of their environmental functions. The functional attributes of a soil microbiome have profound implications for fundamental human needs encompassing food production, environmental protection, and healthcare:
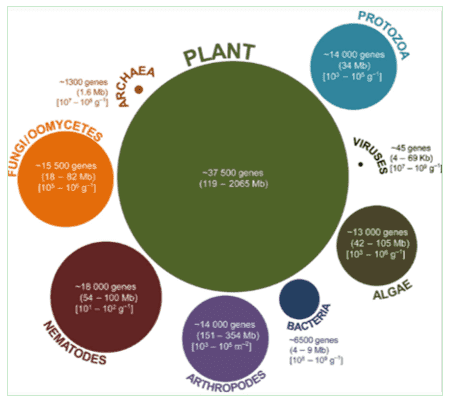
Soil microbiomes hold pivotal roles as primary decomposers within our planetary ecosystem, representing multifaceted environmental and ecological functions. They serve as the engine for biotransformation of vital resources within the soil-plant system. Processes such as decomposition and accumulation of soil organic matter and nitrogen conversion, including biological nitrogen fixation, are invariably linked with microbial activities. Traditional soil microbiology has honed in on these principles, with research focusing on facets such as element transformation rates, nutrient assimilation rates, and correlative associations with relevant soil enzyme activities and functional genes.
Soil microbiomes operate as purification systems for environmental contaminants by effecting biotransformation processes that significantly influence the deposition and form of pollutants within the soil - a key aspect to the soil's function in absorbing pollutants. For organic pollutants, microbial catabolic and co-metabolic processes catalyze transformations or complete breakdowns and mineralization of these pollutants.
Soil microbiomes function as regulators within the context of global environmental change, influencing biogeochemical processes and thereby modulating greenhouse gas emissions and absorption. Soil microbiomes may potentially exacerbate global warming due to their influence on ecosystemic greenhouse gas emissions, inclusive of methane and nitrous oxide. Moreover, they represent a prominent source of nitrous oxide emissions.
Soil microbiomes constitute a critical 'link' orchestrating interplay between above-ground and below-ground interactions within terrestrial ecosystems. They serve as robust drivers of plant diversity and productivity within these ecosystems, directly participating in the delivery of plant nutrients and of soil nutrient cycling.
Soil microbiomes represent a repository of bioactive substances, intimately associated with human health. Beneficial microbes within the soil microbiome can inhibit the propagation and transmission of pathogens affecting humans, plants, and animals, and modifications to land usage can affect human health through alterations to soil microbial diversity. Importantly, soil microbes are a significant resource for secondary metabolite products, many of which have direct applications to healthcare; indeed, numerous natural antibiotics are derived from soil microbes.



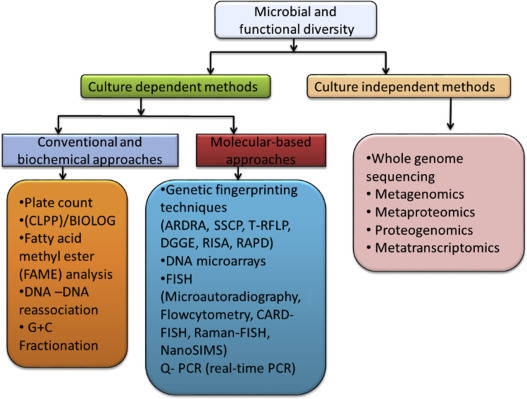
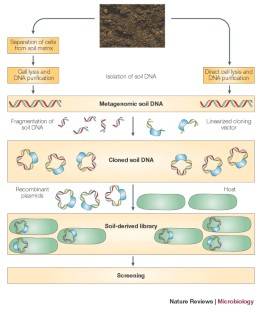 Explore and exploit the genomic diversity of soil microbial communities by metagenomics. (Nature Reviews Microbiology, 470–478, 2005)
Explore and exploit the genomic diversity of soil microbial communities by metagenomics. (Nature Reviews Microbiology, 470–478, 2005)