Phosphorus cycling (PCycle) represents a critical biogeochemical process in which phosphorus (P) transitions between the biosphere, lithosphere, hydrosphere, and atmosphere. As an essential element for life, phosphorus predominantly exists in the form of phosphate and is integral to the structural and functional integrity of vital biological molecules such as DNA, RNA, and ATP. The phosphorus cycle encompasses various stages, including the release of phosphates from rocks through physical and chemical weathering, uptake by plants, and interactions with soil minerals.
1. Phosphorus Release Through Rock Weathering
The weathering of phosphate-rich rocks, driven by physical erosion and chemical processes, introduces both dissolved and particulate phosphorus into the soil. This soil-borne phosphorus, in turn, contributes to the phosphorus pool in aquatic systems via fluvial transport.
2. Phosphorus Uptake and Soil Interactions
Within the soil matrix, phosphate is absorbed by plant roots, facilitating its incorporation into organic matter. However, phosphates can also bind to soil minerals, forming insoluble compounds that are retained within the soil. These interactions can lead to phosphorus immobilization, reducing its bioavailability to plants and other organisms.
3. Aquatic Phosphorus Dynamics
Phosphorus, once dissolved in water, is transported through rivers and eventually reaches lakes and oceans. In aquatic environments, phosphate serves as a vital nutrient for aquatic plants and algae. Nevertheless, excessive phosphorus levels can precipitate eutrophication, driving the proliferation of algal blooms and disrupting aquatic ecosystems.
4. Sedimentation and Phosphorus Burial
Phosphorus in aquatic systems may eventually settle as part of organic and mineral sediments, where it can become buried. Over geological timescales, these sediments may be uplifted, returning to the weathering zone and thus reinitiating the phosphorus cycle.
The Role of Microbes in PCycle
Phosphorus is one of Earth's essential nutrients, and microorganisms play a pivotal role in its cycling. The PCyc database, a comprehensive resource, catalogues 139 gene families associated with 10 distinct phosphorus metabolic processes. These include several phosphorus-cycling genes (PCGs) that have been previously overlooked, such as the pafA gene, which encodes a phosphate-insensitive phosphatase, the ptxABCD gene cluster involved in phosphite metabolism, and the novel aepXVWPS gene set responsible for 2-aminoethylphosphonate transport.
Analysis of Microbial Phosphorus Cycling Genes
To elucidate the role of microbes in phosphorus cycling, metagenomic sequencing is employed to generate non-redundant gene sets from environmental samples. These gene sets are then compared against the PCyc database to identify homologous sequences, allowing for the characterization of PCG families present in the samples. The relative abundance and diversity of these PCGs are visualized, providing insights into the microbial mediation of phosphorus cycling processes.
Table 1. Principal Processes in the Phosphorus Cycle
| PCycle |
Primary Microorganisms |
Environmental Sources |
| Weathering |
Phosphate minerals undergo decomposition during weathering, releasing phosphate ions (PO₄³⁻) into soils and water. |
Promoters of mineral phosphorus dissolution: Mycorrhizal fungi, phosphate-solubilizing bacteria |
| Absorption and Assimilation |
Plants and microorganisms absorb phosphate ions from soil and incorporate them into organic molecules. |
Mycorrhizal fungi, Pseudomonas spp., Rhizobium |
| Decomposition |
After the death of plants and animals, organic phosphorus compounds are decomposed into inorganic phosphate, re-entering the soil. |
Saprotrophic bacteria and fungi: Bacillus spp., Aspergillus spp. |
| Mineralization |
Organic phosphorus compounds are decomposed by microorganisms into inorganic phosphate. |
Phosphomineralizing bacteria: Bacillus spp., Pseudomonas spp. |
| Sedimentation |
Phosphate precipitates from aquatic systems, forming sediments that are stored in sedimentary rocks over long periods. |
This process is primarily physical and chemical; some microorganisms can influence sediment formation and dissolution. |
| Recycling |
Phosphate ions in aquatic environments are absorbed by phytoplankton and other aquatic organisms, entering the aquatic food web. |
Phytoplankton, certain aquatic bacteria |
Table 2. Comparative Analysis of Biogeochemical Cycles (Nitrogen Cycle, Sulfur Cycle, Phosphorus Cycle)
| NCycle |
SCycle |
PCycle |
| Environmental Sources |
Atmosphere, soil |
Phosphate minerals, soil |
| Processes |
Nitrogen fixation, nitrification, assimilation, denitrification |
Mineralization, sulfur oxidation, assimilation, dissimilatory sulfate reduction |
| Primary Microorganisms |
Nitrogen-fixing bacteria, nitrifying bacteria, denitrifying bacteria |
Sulfur-oxidizing bacteria, sulfate-reducing bacteria |
| Analytical Strategy |
Similar |
Similar |
Analytical Strategies for PCycle in Ecosystems
PCycle is a critical biogeochemical process involving the movement, transformation, and recycling of phosphorus within ecosystems and the environment. Phosphorus is one of the essential nutrients required for plant growth and reproduction, playing a pivotal role in various biological functions.
In natural ecosystems, phosphorus primarily exists as phosphate, which is released from rocks and soils through weathering and leaching processes. This phosphate is transported into rivers, eventually reaching oceans, where it deposits on the seafloor. Geological processes may later expose these deposits, reintroducing phosphorus into the cycle. The phosphorus cycle encompasses both terrestrial and aquatic ecosystems, with distinct dynamics in each.
Human activities, such as the extraction of phosphate rock, the production and use of phosphorus-based fertilizers and pesticides, and the discharge of phosphorus-containing industrial and domestic wastewater, have significantly altered the natural phosphorus cycle. These interventions have led to changes in phosphorus availability, often resulting in environmental issues such as eutrophication in aquatic systems.
The analysis of phosphorus cycling within microbial communities, particularly through metagenomic approaches, has become a focal point in contemporary research. The primary strategies for analyzing microbial PCycle data include the following:
Table 3. Comparative Analysis of PCycle Analytical Strategies
| Aspect |
Analysis Strategy One |
Analysis Strategy Two |
Analysis Strategy Three |
| Analysis Focus |
Combination of different databases |
Phenotypic information mining |
MAGs analysis |
| Core Objective |
Identification of target functional genes and species in the PCycle |
Core factors influencing various processes of the PCycle |
Acquisition of complete and accurate species and functional information |
| Analytical Methods |
Joint analysis with the EggNOG database; construction of PCycle functional gene co-occurrence networks; integration with existing research reports |
Regression analysis of environmental factors |
Structural analysis of biological genes from MAGs |
Analytical Strategy 1: Integration of PCycle with Other Database Annotations
The integration of PCycle data with annotations from other databases is a crucial approach in understanding the functional roles of microbial communities in phosphorus cycling. The following outlines the methodological steps involved in this analytical strategy:
Microbial Community Profiling via Metagenomic Sequencing:
Comprehensive metagenomic sequencing is employed to obtain a detailed profile of the microbial community, including species distribution and the identification of differential markers and genes across various comparative groups. This step provides a foundational understanding of the microbial ecosystem and its dynamics.
Identification of PCGs:
Utilizing the results from PCycle analysis, researchers can identify key PCGs. A co-occurrence network analysis of these PCGs is conducted to pinpoint core functional genes. This analysis facilitates the association of critical functional genes with specific target species within the microbial community.
Functional Gene Classification Using the EggNOG Database:
The results from the PCycle analysis are further integrated with the EggNOG database, which provides a comprehensive framework for protein function classification. This integration allows for the association of target genes with their respective protein functions. For a detailed discussion on the strategies and methodologies employed in EggNOG database analysis, refer to the article titled "How to Identify the Functional Roles of Target Genes in Microbial Communities."
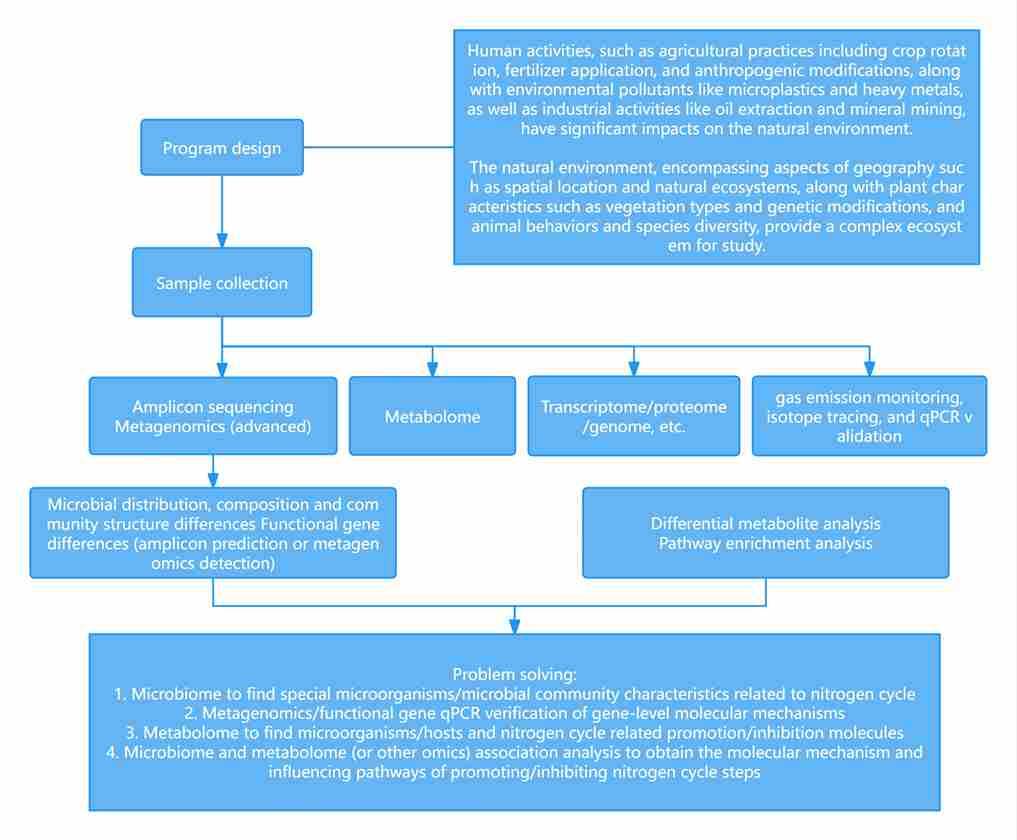
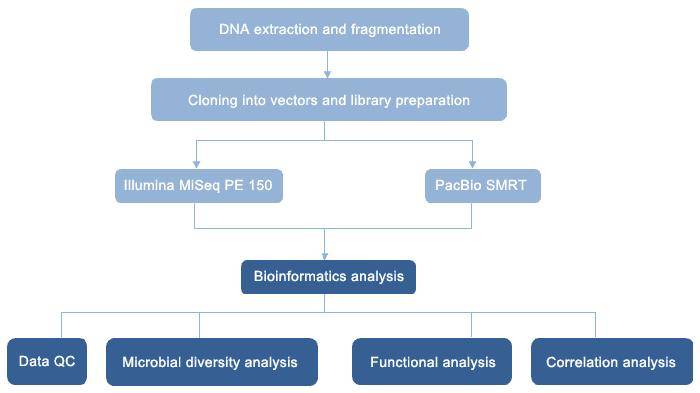 Figure 1 Metagenomics Analysis Workflow
Figure 1 Metagenomics Analysis Workflow
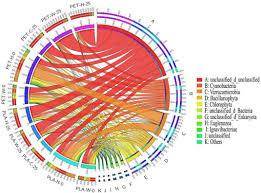 Figure 2 Phosphorus cycle (PCycle) analysis Circos diagram Xiaojun Song (et al,. 2023)
Figure 2 Phosphorus cycle (PCycle) analysis Circos diagram Xiaojun Song (et al,. 2023)
Analytical Strategy 2: Identification of Key Environmental Factors Influencing PCycle
PCycle is a critical ecological process influenced by a myriad of environmental factors. The identification and analysis of these factors are paramount to understanding and optimizing the phosphorus cycle within ecosystems. The following steps outline this analytical strategy:
Targeted Identification of PCGs:
The PCycle analysis is employed to identify target PCGs. This step is essential for pinpointing the genes that play crucial roles in various stages of the phosphorus cycle within microbial communities.
Correlation of Environmental Factors with PCGs:
The functional genes identified through PCycle analysis are then correlated with environmental factor data. This correlation analysis is designed to determine the key environmental variables—such as soil physicochemical properties—that significantly influence the phosphorus cycling process. These factors could include soil pH, moisture content, organic matter, and nutrient availability, all of which may critically affect the functionality of the identified PCGs.
Analytical Strategy 3: Metagenome-Assembled Genomes (MAGs) for Comprehensive Gene Collection
MAGs analysis represents a powerful approach for reconstructing relatively complete genomic sequences from complex microbial communities. This strategy is particularly advantageous for capturing the full spectrum of PCGs within diverse ecosystems. The methodology is as follows:
Application of MAGs Analysis for Genome Assembly:
MAGs analysis is utilized to assemble genome sequences from metagenomic data, providing a more complete and accurate genomic context for the target PCGs. This method enables the identification of comprehensive gene sets involved in phosphorus cycling, which might otherwise be fragmented in traditional metagenomic analyses.
Enhanced Applicability in Soil Ecosystem Studies:
The PCycle analysis is predominantly applied in the study of soil ecosystems, where phosphorus cycling plays a vital role. The integration of MAGs analysis within this context is particularly suitable for studies involving complex community data, as it offers a more holistic view of the microbial processes at play. This approach, as implemented in the newly updated MAGs analysis pipeline, is expected to yield more accurate and predictable results in soil microecology research, particularly concerning phosphorus cycling.
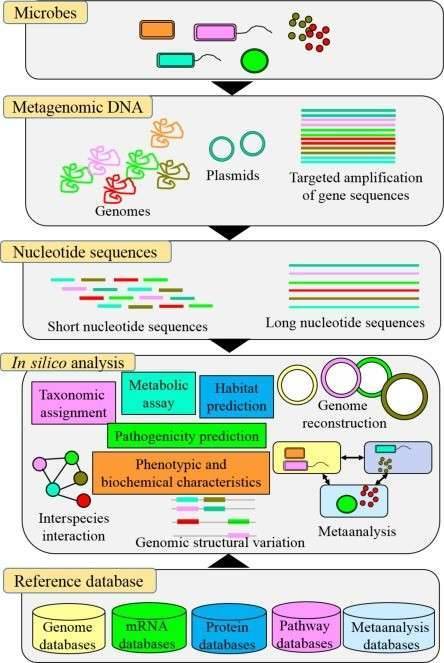
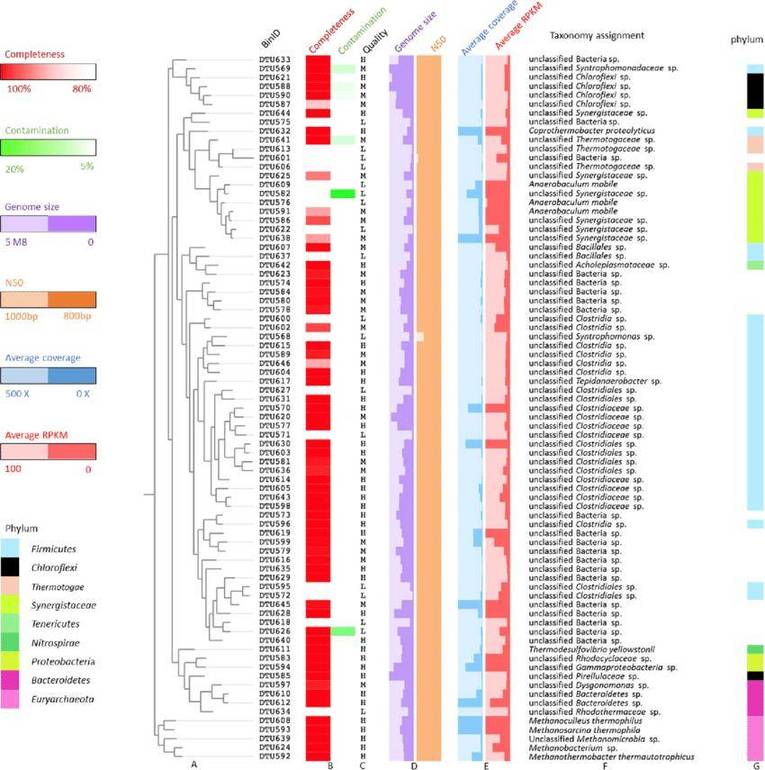 Figure 3 Metagenome assembled genomes (MAGs)
Figure 3 Metagenome assembled genomes (MAGs)
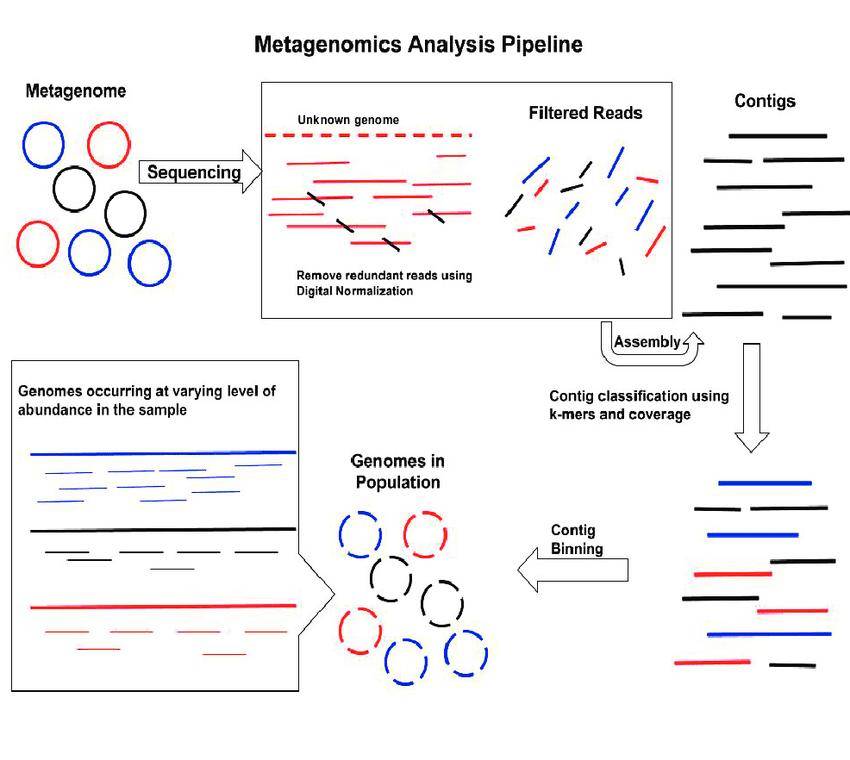 Figure 4 Metagenomic MAGs pipeline (Jay Ghurye et al,. 2016)
Figure 4 Metagenomic MAGs pipeline (Jay Ghurye et al,. 2016)
Classic Case Study of PCycle Analysis
Title: Microbial Phosphorus-Cycling Genes in Soil Under Global Change
Journal: Wiley (Impact Factor: 10.4)
Publication Date: 2024
Introduction
The Qinghai-Tibet Plateau, a region highly sensitive to climate change, has experienced significant alterations in temperature and precipitation patterns. These environmental shifts have profoundly impacted the phosphorus cycling system within alpine meadow soils. However, the intricate interactions and cascading effects of these environmental factors—particularly warming and changes in precipitation—on key microbial "extracellular" and "intracellular" PCGs remain largely unexplored. This study aims to elucidate the effects of warming and precipitation changes on soil PCGs and phosphorus transformation processes, while also investigating the roles of these environmental factors on microbial dynamics within the soil microecosystem.
Main Findings
Assembly and Binning of Metagenomic Data:
MAGs were derived from metagenomic sequencing data through assembly and binning processes. The gene sets obtained from these MAGs were then compared with the PCycDB database, a specialized repository for phosphorus cycling genes, to classify the PCGs. This comprehensive analysis revealed 120 genes associated with soil phosphorus cycling, covering 10 distinct phosphorus cycling pathways.
In all soil samples, PCGs were predominantly enriched in pathways related to purine metabolism, transport proteins, and pyrimidine metabolism. These pathways exhibited the highest abundance of PCGs, underscoring their critical roles in the soil phosphorus cycle.
The study uncovered a negative correlation between the abundance of PCGs and individual climate factors—namely increased precipitation, decreased precipitation, and isolated warming. Conversely, under the combined influence of warming and precipitation changes, an increase in the abundance of PCGs was observed. This finding suggests that the synergistic effects of these environmental factors may enhance the microbial capacity for phosphorus cycling in soil ecosystems.
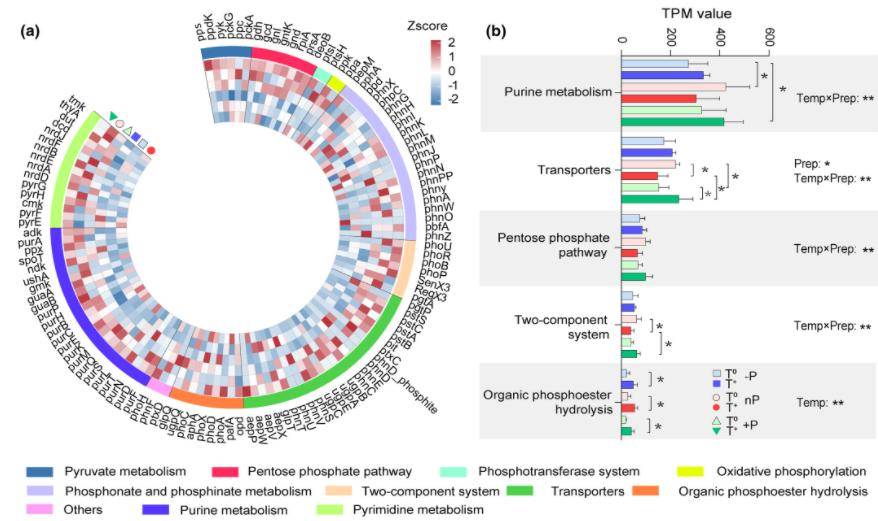 Figure 5. Relative abundances of 120 microbial PCycle functional genes in soil samples. Asterisks denote significant impacts of warming and precipitation environmental factors on the relative abundances of these PCycle functional genes.
Figure 5. Relative abundances of 120 microbial PCycle functional genes in soil samples. Asterisks denote significant impacts of warming and precipitation environmental factors on the relative abundances of these PCycle functional genes.
Metagenomic sequencing
Metagenomic sequencing was employed to identify and classify PCycle functional genes from soil samples. The classification of these genes was conducted using metagenomic data, which were analyzed in conjunction with environmental factor datasets to determine the main influences on various stages of the phosphorus cycling process. This approach enabled the identification of specific genes that play crucial roles in the transformation and circulation of phosphorus within the soil.
The analysis identified 54 distinct PCGs that exhibited significant correlations with soil phosphorus components. This finding underscores the critical role of these genes in regulating phosphorus transformation and cycling within alpine meadow soils. The significant correlation between these PCGs and soil phosphorus components indicates that these functional genes may influence the bioavailability and mobility of phosphorus in these environments. The implications of this are profound, as they suggest that shifts in environmental factors, such as temperature and precipitation, could have direct effects on the functionality of these genes, thereby altering the phosphorus cycle in the soil.
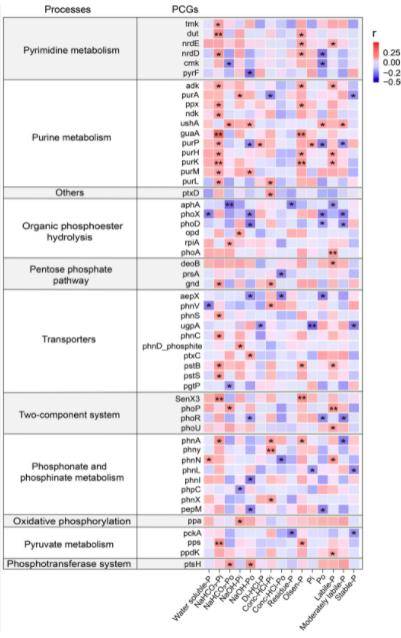 Figure 6. Correlation between PCycle functional genes and soil phosphorus components under the combined effects of warming and precipitation.
Figure 6. Correlation between PCycle functional genes and soil phosphorus components under the combined effects of warming and precipitation.
Analysis of Microbial Community Composition and Dynamics Through Metagenomic Sequencing
Metagenomic sequencing was employed to investigate the overall species distribution and trends within microbial communities. The analysis focused on assembling MAGs from the metagenomic data to obtain high-quality microbial genomes. A total of 16 MAGs were successfully reconstructed, each exhibiting greater than 80% completeness and less than 10% contamination. These MAGs were taxonomically classified using the Genome Taxonomy Database Toolkit (GTDB-Tk), enabling a robust phylogenetic analysis of the microbial taxa present in the samples.
The phylogenetic classification of the 16 MAGs revealed their affiliation with various bacterial taxa, including Streptococcus, Cytophagales, Flavobacteriaceae, and Alphaproteobacteria. These taxa represent a significant portion of the microbial community within the studied ecosystem under baseline environmental conditions. However, under the combined effects of warming and altered precipitation, a notable shift was observed, with the MAGs predominantly classified as Acidobacteriota. This shift suggests that environmental changes exert selective pressures on microbial communities, favoring taxa that are better adapted to altered conditions.
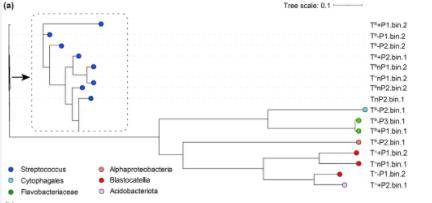 Figure 7. Taxonomic classification of 16 MAGs based on GTDB-Tk taxonomy.
Figure 7. Taxonomic classification of 16 MAGs based on GTDB-Tk taxonomy.
Summary
Phosphorus cycling is a multifaceted ecological process encompassing geological, chemical, and biological dimensions. Anthropogenic disturbances to the phosphorus cycle can lead to significant environmental issues, necessitating the scientific management and judicious use of phosphorus resources to maintain ecosystem balance and ensure sustainable development. PCycle is predominantly observed within soils, aquatic systems, and biological organisms. Analysis of SCycle is particularly crucial for soil microbiome research. The following summary encapsulates the primary aspects of PCycle research based on the preceding analyses:
Annotation of Phosphorus Cycling Functional Genes: The analysis of PCycle provides annotated information on PCycle functional genes across different habitats.
Functional Information Acquisition: Integrating PCycle analysis with the EggNOG database yields specific functional details of target genes.
Environmental Factor Assessment: By combining PCycle functional genes with environmental factor data, the key environmental variables influencing entire habitats can be inferred.
CD Genomics has developed advanced metagenomic analysis workflows and MAGs analysis processes, facilitating the execution of specialized and customized analyses for metagenomic data exploration.


 Figure 1 Metagenomics Analysis Workflow
Figure 1 Metagenomics Analysis Workflow Figure 2 Phosphorus cycle (PCycle) analysis Circos diagram Xiaojun Song (et al,. 2023)
Figure 2 Phosphorus cycle (PCycle) analysis Circos diagram Xiaojun Song (et al,. 2023) Figure 3 Metagenome assembled genomes (MAGs)
Figure 3 Metagenome assembled genomes (MAGs) Figure 4 Metagenomic MAGs pipeline (Jay Ghurye et al,. 2016)
Figure 4 Metagenomic MAGs pipeline (Jay Ghurye et al,. 2016) Figure 5. Relative abundances of 120 microbial PCycle functional genes in soil samples. Asterisks denote significant impacts of warming and precipitation environmental factors on the relative abundances of these PCycle functional genes.
Figure 5. Relative abundances of 120 microbial PCycle functional genes in soil samples. Asterisks denote significant impacts of warming and precipitation environmental factors on the relative abundances of these PCycle functional genes. Figure 6. Correlation between PCycle functional genes and soil phosphorus components under the combined effects of warming and precipitation.
Figure 6. Correlation between PCycle functional genes and soil phosphorus components under the combined effects of warming and precipitation. Figure 7. Taxonomic classification of 16 MAGs based on GTDB-Tk taxonomy.
Figure 7. Taxonomic classification of 16 MAGs based on GTDB-Tk taxonomy.