Wide Applications of MeRIP-seq in Biomedicine, Especially in Diseases Diagnosis
Methylated RNA immunoprecipitation sequencing (MeRIP-seq) is predominantly employed to investigate the methylation modifications of RNA and finds extensive utilization in the biomedical realm. It has the capacity to disclose the dynamic alterations in RNA methylation during the onset and progression of numerous diseases, such as tumors and nervous system disorders, thereby facilitating the discovery of potential biomarkers. Additionally, it can be harnessed to explore the regulatory mechanisms of RNA methylation in normal physiological processes, including cell differentiation and development, thus contributing to the elucidation of the essence of life activities. Simultaneously, in the domain of drug research and development, it can furnish crucial information for assessing drug efficacy and probing drug targets.
Study on Disease Mechanism
Epigenetic regulation: MeRIP-seq can be utilized to identify RNA methylation sites on disease-related genes and unravel the role of epigenetic regulation in the pathogenesis of diseases. For instance, in cancer research, it has been observed that alterations in the m6A RNA methylation levels of certain genes are intimately associated with tumor proliferation, invasion, and metastasis.
Post-transcriptional regulation: By detecting the impacts of RNA methylation on mRNA stability, splicing, and translation, MeRIP-seq is conducive to clarifying the regulatory mechanisms of gene expression in diseases. In the study of neurodevelopment-related diseases, it has been discovered that the m6A RNA methylation modification of some long-chain noncoding RNAs (lncRNAs) can modulate their interaction with miRNAs, thereby influencing the differentiation and development of nerve cells.
Targets for Disease Treatment
Drug research and development: The development of specific inhibitors or activators for RNA methylation-related enzymes and proteins, such as methyltransferases and demethylases, represents a novel disease treatment strategy. MeRIP-seq can assist in determining the targets of these drugs and enhance the efficiency and relevance of drug research and development. In the study of osteosarcoma, it was found that ALKBH5 is an important m6A RNA demethylase, and its overexpression is closely correlated with the growth and metastasis of osteosarcoma. Through MeRIP-seq, the downstream target genes USP22 and RNF40 were identified, providing a new target for the treatment of osteosarcoma.
Gene therapy: Utilizing RNA editing technologies, such as the CRISPR-Cas9 system, to edit disease-related genes and rectify abnormal methylation statuses of genes is a potential disease treatment approach. MeRIP-seq can help to identify the gene sites that require editing and improve the efficacy of gene therapy. In the study of some hereditary diseases, it has been found that the m6A RNA methylation modification of genes leads to abnormal gene expression, and it is anticipated that the treatment of diseases can be achieved by employing gene editing technology to repair methylation defects.
Diagnostic Markers of Diseases
Biomarker discovery: By comparing the RNA methylation profiles of disease patients and healthy individuals, MeRIP-seq can identify disease-specific methylation markers. These markers can serve as biomarkers for the diagnosis, classification, and prognosis of diseases, thereby enhancing the diagnostic accuracy and therapeutic effectiveness of diseases. In the study of gastric cancer, MeRIP-seq detected site-specific m6A RNA modifications of PSMA3-AS1 and MIR22HG, which were found to be related to the apoptosis and stemness of gastric cancer stem cells, presenting potential targets for the diagnosis and treatment of gastric cancer.
Early diagnosis: Since RNA methylation may change in the early stages of a disease, MeRIP-seq is beneficial for the early diagnosis of the disease. In the study of Alzheimer's disease, it was discovered that the m6A RNA methylation level of some neuron-specific genes significantly decreased in the early stage of the disease, providing important clues for early diagnosis.
Examples of MeRIP-seq in Disease Diagnosis
MeRIP-seq technology is a crucial method for studying RNA methylation and is widely applied in disease diagnosis. The following are some illustrative examples of the application of MeRIP-seq technology in disease diagnosis:
Cancer diseases
Gastric cancer: In the research on gastric cancer, MeRIP-seq technology revealed that the site-specific m6A RNA modification of two long-chain noncoding RNAs, PSMA3-AS1 and MIR22HG, is closely related to the apoptosis and stemness of gastric cancer stem cells. Methylated PSMA3-AS1 and MIR22HG can inhibit the apoptosis of gastric cancer stem cells and promote their stemness, thereby facilitating the occurrence and development of tumors. These findings offer a new target for the diagnosis and treatment of gastric cancer.
Osteosarcoma: For osteosarcoma, MeRIP-seq technology disclosed that the RNA demethylase ALKBH5 affects the growth and progression of osteosarcoma by regulating the m6A RNA methylation levels of the histone ubiquitination enzyme USP22 and the ubiquitin ligase RNF40. The amplification and overexpression of ALKBH5 are associated with the poor prognosis of osteosarcoma, and inhibiting the activity of ALKBH5 can be regarded as a potential treatment strategy for osteosarcoma.
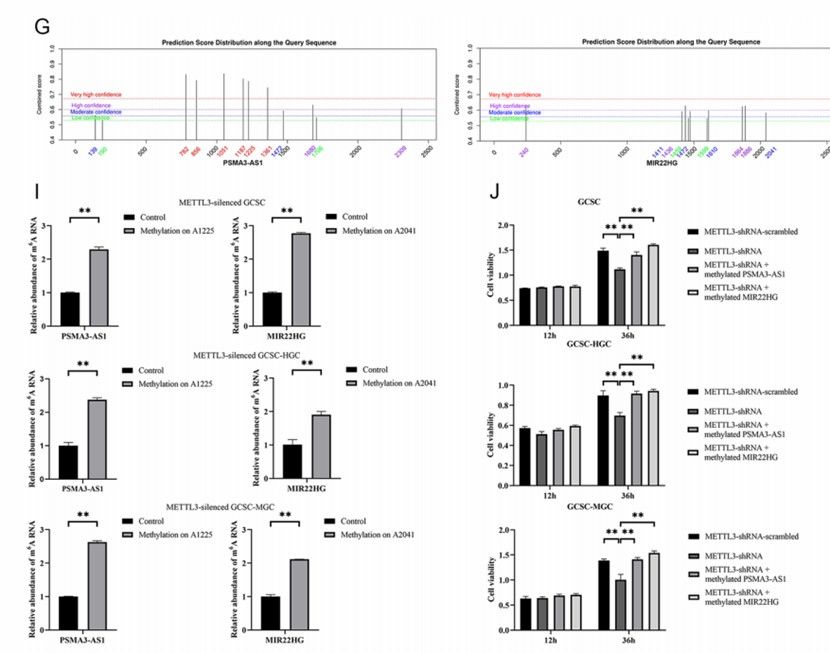 Roles of methylated lncRNAs in GCSCs (Ci et al., 2024)
Roles of methylated lncRNAs in GCSCs (Ci et al., 2024)
Nervous System Diseases
Alzheimer's Harmo's disease: In the study of Alzheimer's disease, MeRIP-seq technology detected that the m6A RNA methylation level of some neuron-specific genes significantly decreased in the early stage of the disease. Abnormal methylation of these genes may impact the function and survival of neurons, leading to the death of nerve cells and cognitive dysfunction. By detecting these methylation markers, evidence for the early diagnosis of Alzheimer's disease can be provided.
Parkinson's disease: MeRIP-seq technology also identified that the m6A RNA methylation level of some genes related to neurotransmitter release and dopamine metabolism changed in the brains of Parkinson's patients. These alterations may affect the balance of neurotransmitters and the function of dopaminergic neurons, thus resulting in the symptoms of Parkinson's disease.
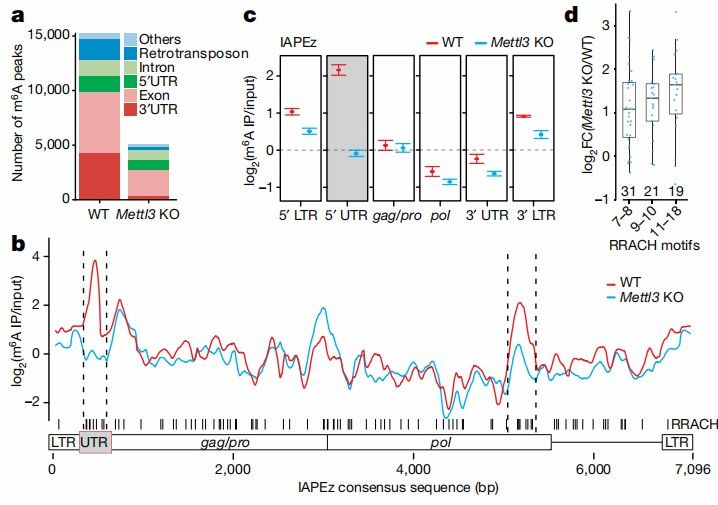 M6A methylation of IAP mRNAs (Tomasz et al., 2020)
M6A methylation of IAP mRNAs (Tomasz et al., 2020)
Cardiovascular Diseases
Myocardial infarction: In the study of myocardial infarction, MeRIP-seq technology found that the m6A RNA methylation level of some genes related to myocardial apoptosis and fibrosis changed in the late stage of myocardial infarction. These changes may influence the repair and regeneration ability of myocardial cells and lead to the deterioration of myocardial function. By detecting these methylation markers, the degree of injury and prognosis of myocardial infarction can be evaluated.
Coronary heart disease: By analyzing cardiovascular tissue samples from patients with coronary heart disease and healthy controls using MeRIP-seq technology, differences in the m6A RNA methylation levels of genes related to myocardial cell metabolism, inflammatory reaction, and vascular smooth muscle cell function were detected. These differential methylation sites can serve as diagnostic markers of coronary heart disease, facilitating early diagnosis and risk assessment.
Heart failure: In the study of heart failure, MeRIP-seq technology can detect the M {6} A methylation status of genes related to myocardial remodeling, cardiac fibrosis, and neuroendocrine regulation. For example, it was found that the m6A RNA methylation modification of some lncRNAs is related to the severity of heart failure, which can be used as a diagnostic and prognostic index of heart failure.
Infectious diseases
Virus infection: MeRIP-seq technology can be employed to investigate the effect of virus infection on RNA methylation in host cells. In hepatitis B virus infection, MeRIP-seq technology discovered that the m6A RNA methylation level of the hepatitis B virus gene is closely related to virus replication and transcription. By detecting the changes in RNA methylation in host cells, the interaction between the virus and host cells can be understood, providing novel ideas for the diagnosis and treatment of virus infection.
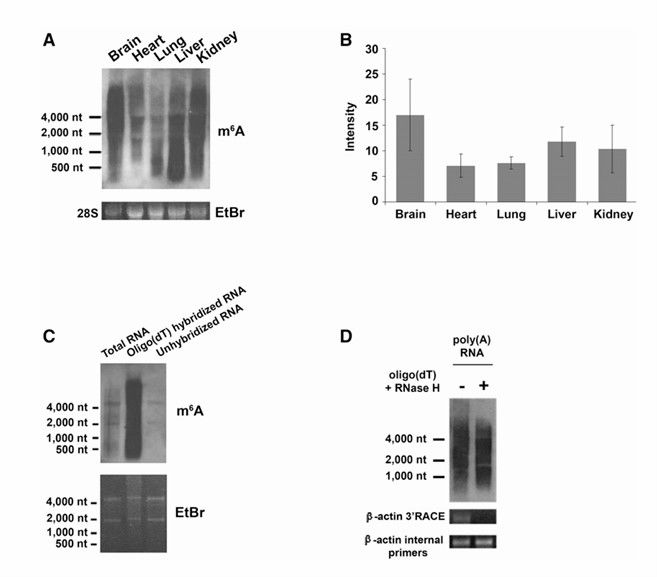 Different levels of m6A RNA in organs (Kate et al., 2012)
Different levels of m6A RNA in organs (Kate et al., 2012)
Digestive System Diseases
Inflammatory bowel disease: MeRIP-seq technology can be used to study the changes in RNA methylation in the intestinal tissue of patients with inflammatory bowel disease. It was found that the m6A RNA methylation level of genes related to intestinal mucosal immunity, inflammatory response, and apoptosis changed in inflammatory bowel disease, which may be involved in the pathogenesis of the disease, providing a new target for the diagnosis and treatment of inflammatory bowel disease.
Liver Diseases
Hepatitis B: For hepatitis B, MeRIP-seq technology can analyze the changes in RNA methylation in host cells after hepatitis B virus infects hepatocytes. It was found that the m6A RNA methylation level of some genes related to virus replication, immune response, and hepatocyte injury is closely related to the infection status and disease progression of hepatitis B virus, which can be used as a monitoring index for the diagnosis and treatment of hepatitis B.
Hepatitis C: In the study of hepatitis C, MeRIP-seq technology is helpful in revealing the interaction mechanism between hepatitis C virus and host cells and identifying the m6A RNA methylation changes of genes related to virus infection, inflammatory reaction, and hepatocellular carcinoma, thus providing evidence for the diagnosis and treatment of hepatitis C.
Blood System Diseases
Leukemia: MeRIP-seq technology can be used to study the characteristics of RNA methylation in leukemia cells, and it was found that the m6A RNA methylation level of genes related to the proliferation, differentiation, and apoptosis of leukemia cells has changed. These changes can serve as diagnostic markers of leukemia, aiding in the differentiation of different types of leukemia and providing new strategies for the treatment of leukemia.
Lymphoma: In the study of lymphoma, MeRIP-seq technology can detect the m6A RNA methylation status of genes related to the immunophenotype of lymphoma cells, tumor cell proliferation, and drug resistance, providing important information for the diagnosis and treatment of lymphoma.
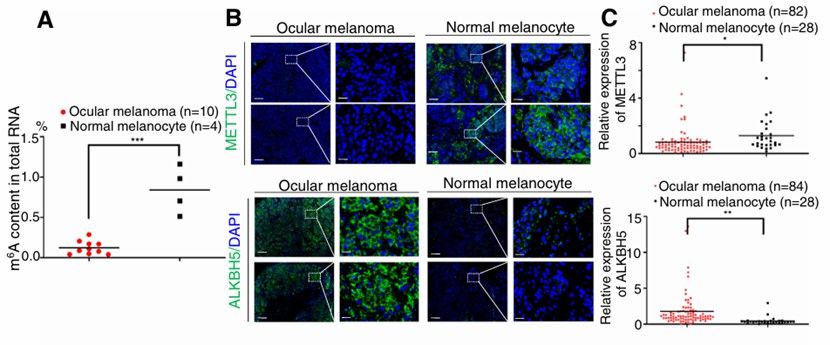 Decreased level of m6A RNA in ocular melanoma (Jia et al., 2019)
Decreased level of m6A RNA in ocular melanoma (Jia et al., 2019)
Respiratory Diseases
Asthma: By analyzing airway epithelial cell samples from asthmatic patients and healthy controls using MeRIP-seq technology, differences in the m6A RNA methylation levels of genes related to airway inflammation, immune response, and mucus secretion were detected. These differential methylation sites can be used as diagnostic markers of asthma, facilitating the understanding of the pathogenesis of asthma and the formulation of personalized treatment plans.
Chronic obstructive pulmonary disease: In the study of COPD, MeRIP-seq technology can detect the m6A RNA methylation status of genes related to pulmonary inflammation, oxidative stress, and apoptosis and identify some methylation markers related to the severity of COPD, providing a basis for the diagnosis and prognosis evaluation of COPD.
Future Prospects of MeRIP-seq in Diseases
Technical improvement: With the continuous advancement of technology, the detection sensitivity, specificity, and accuracy of MeRIP-seq will be further enhanced. For example, improving the quality and specificity of antibodies, optimizing the experimental operation process, and increasing the depth and quality of sequencing will enable better detection of low-abundance methylation sites and complex methylation patterns.
Multiomics integration: MeRIP-seq technology will be integrated with other omics technologies, such as transcriptomics, proteomics, and metabolomics, to realize the comprehensive analysis of multiomics data. This will assist in a more comprehensive understanding of the pathogenesis of diseases, the discovery of more disease-related biomarkers, and the provision of more information for the diagnosis and treatment of diseases.
Early marker discovery: MeRIP-seq technology can detect changes in RNA methylation in the early stages of a disease, offering the possibility of early diagnosis. By analyzing samples from high-risk groups or individuals with prodromal symptoms, it is expected that early diagnostic markers can be identified, enabling early intervention and treatment of diseases and improving patient survival rates and quality of life.
Sensitive detection: This technique exhibits high sensitivity and specificity and can detect small amounts of RNA samples, which is suitable for the characteristics of small samples in early diseases. For example, MeRIP-seq technology can play a significant role in the early diagnosis of some rare diseases or tumors.
Study on disease heterogeneity: Many diseases display high heterogeneity. MeRIP-seq technology can deeply investigate the heterogeneity mechanism of diseases, identify differences among different subtypes of diseases, and provide support for the accurate treatment of diseases.
Prognostic value of methylation markers: Specific RNA methylation markers are closely related to the prognosis of diseases, and MeRIP-seq technology can evaluate the prognosis of diseases by detecting changes in these markers. In cancer research, for instance, the m6A RNA methylation level of some genes is associated with tumor recurrence, metastasis, and patient survival rate, which can serve as an important prognostic indicator for cancer.
Clinical transformation: The development of MeRIP-seq technology will promote its application transformation in clinical diagnosis. An increasing number of research institutions and clinical laboratories will conduct application research on MeRIP-seq technology, establish standardized detection procedures and quality control systems, and provide reliable technical support for clinical diagnosis.
Personalized medical care: The disease diagnosis method based on MeRIP-seq technology can offer strong support for personalized medical care. By analyzing the RNA methylation characteristics of individual patients, personalized treatment plans can be formulated to enhance the pertinence and effectiveness of treatment and reduce drug adverse reactions.
Disease prevention: Understanding the changes in RNA methylation during the occurrence and development of diseases is beneficial for formulating targeted disease prevention strategies. For example, by intervening in the RNA methylation process related to diseases, the occurrence and development of diseases can be prevented, and the incidence of diseases can be reduced.
In summary, MeRIP-seq technology holds significant application value and broad application prospects in disease diagnosis. With the continuous development and improvement of technology, it will provide more accurate and effective methods for the early diagnosis, classification, prognosis evaluation, and personalized treatment of diseases.
References:
- Tomasz C, Emeline R, Aurelie T, et al. m6A RNA methylation regulates the fate of endogenous retroviruses, 2.13 (2021),Nature, https://doi.org/10.1038/s41586-020-03135-1.
- Ci Y, Zhang Y, Zhang XB. Methylated lncRNAs suppress apoptosis of gastric cancer stem cells via the lncRNAs-miRNA/protein axis, (2024) 29: 51, Cellular & Molecular Biology Letters, https://doi.org/10.1186/s11658-024-00568-8.
- Yang WL, Zhao YL, Yang YG. Dynamic RNA methylation modifications and their regulatory role in mammalian development and disease, 5.31 (2024): 2084-2104, Science China Life Sciences, https://doi.org/10.1007/s11427-023-2526-2.
- Pooja Y, Panneerdoss S, Daisy M, et al. M6A RNA Methylation Regulates Histone Ubiquitination to Support Cancer Growth and Progression, (2022): 1872-1889, Cancer Res, DOI: 10.1158/0008-5472.CAN-21-2106.
- Yuri M, Mark H. Methods for RNA Modification Mapping Using Deep Sequencing: Established and New Emerging Technologies, 1.9, (2019) 10:35, Genes, DOI:10.3390/genes10010035.
- Chao XH, Guo LJ, Ye CT, et al. ALKBH5 regulates chicken adipogenesis by mediating LCAT mRNA stability depending on m6 A modiffcation, (2024) 25:634, BMC Genomics, https://doi.org/10.1186/s12864-024-10537-2.
- Jia RB, Chai PW, Wang SZ, et al. M6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation, (2019) 18:161, Molecular Cancer, https://doi.org/10.1186/s12943-019-1088-x.
- Yin HL, Zhang X, Yang PY, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming, (2021) 12:1394, Nature Communications, https://doi.org/10.1038/s41467-021-21514-8.
- Shen CQ, Xuan BQ, Yan TT, et al. M6A-dependent glycolysis enhances colorectal cancer progression, (2020) 19:72, Molecular Cancer, https://doi.org/10.1186/s12943-020-01190-w.
- An YY, Duan H. The role of m6A RNA methylation in cancer metabolism, (2022) 21:14, Molecular Cancer, https://doi.org/10.1186/s12943-022-01500-4.
- Kade D, Yogesh S, Paul Z, et al. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 30 UTRs and near Stop Codons, 6.22, (2012), 1635-1646, Cell, DOI 10.1016/j.cell.2012.05.003.
- Wang YB, Chen Y, Xiao HF, et al, METTL3-mediated m6A modification increases Hspa1a stability to inhibit osteoblast aging, 3.27, (2024), Official journal of CDDpress, https://doi.org/10.1038/s41420-024-01925-4.
! For research purposes only, not intended for clinical
diagnosis, treatment, or individual health assessments.
 Roles of methylated lncRNAs in GCSCs (Ci et al., 2024)
Roles of methylated lncRNAs in GCSCs (Ci et al., 2024) M6A methylation of IAP mRNAs (Tomasz et al., 2020)
M6A methylation of IAP mRNAs (Tomasz et al., 2020) Different levels of m6A RNA in organs (Kate et al., 2012)
Different levels of m6A RNA in organs (Kate et al., 2012) Decreased level of m6A RNA in ocular melanoma (Jia et al., 2019)
Decreased level of m6A RNA in ocular melanoma (Jia et al., 2019)




