Pseudouridine (Ψ) is the most abundant RNA modification found in non-coding RNAs (ncRNAs) and was one of the first modifications discovered in RNA hydrolysates. Recent advancements in high-throughput sequencing have revealed that Ψ is also prevalent in messenger RNAs (mRNAs), playing crucial roles in RNA stability, translation efficiency, and gene regulation. Due to its unique structural properties, Ψ influences processes such as translation fidelity and alternative splicing, with potential links to diseases like cancer and immune responses. PA-Ψ-seq (photo-crosslinking-assisted Ψ sequencing, with "PA" standing for "Primer-anchored") from CD Genomics is a cutting-edge technology that enables precise and high-resolution detection of pseudouridine modifications, offering comprehensive insights into the transcriptome-wide distribution of Ψ.
PA-Ψ-seq is distinct from other pseudouridine detection technologies, such as Pseudo-seq and Ψ-seq, primarily due to its unique primer-anchored strategy. This innovative approach significantly enhances the resolution and accuracy of pseudouridine detection. The specific primer design and sequence alignment analysis employed in PA-Ψ-seq provide advantages in detection efficiency and data quality compared to other methods.
Specific PA-Ψ-seq Approaches We Offer
Our PA-Ψ-seq services provide:
- Single-nucleotide resolution detection of Ψ modifications
- Comprehensive mapping of Ψ modifications across the transcriptome
- Differential Ψ modification analysis in disease contexts, such as cancer
Methods
The PA-Ψ-seq method involves several critical steps designed to accurately capture and analyze pseudouridine modifications in RNA:
- Antibody Binding and UV Crosslinking: Total poly(A) RNA is combined with an anti-pseudouridine (anti-Ψ) antibody and RNasin, allowing specific binding to Ψ-modified RNA. This mixture is then subjected to UV crosslinking to covalently link the RNA to the antibody, enhancing specificity.
- RNA Digestion and Immunoprecipitation: Following crosslinking, RNase T1 is added to digest unmodified RNA, ensuring only Ψ-modified RNA remains bound. Protein G magnetic beads are then introduced to pull down the RNA-antibody complexes, which are subsequently eluted using Proteinase K.
- RNA Purification and End Repair: The eluted Ψ-modified RNA is purified through phenol-chloroform extraction and ethanol precipitation. The purified RNA undergoes treatment with Calf Intestinal Phosphatase (CIP) to remove 5' phosphate groups, followed by T4 Polynucleotide Kinase (T4-PNK) to phosphorylate the 5' ends.
- Library Construction and Sequencing: The EPI miRNA Library Kit is utilized to construct the sequencing library, which involves adding specific adapters, performing reverse transcription to synthesize cDNA, and amplifying the cDNA through PCR. Finally, the prepared library is subjected to high-throughput sequencing to generate raw data for analysis.
The technical principle is illustrated in the diagram below:
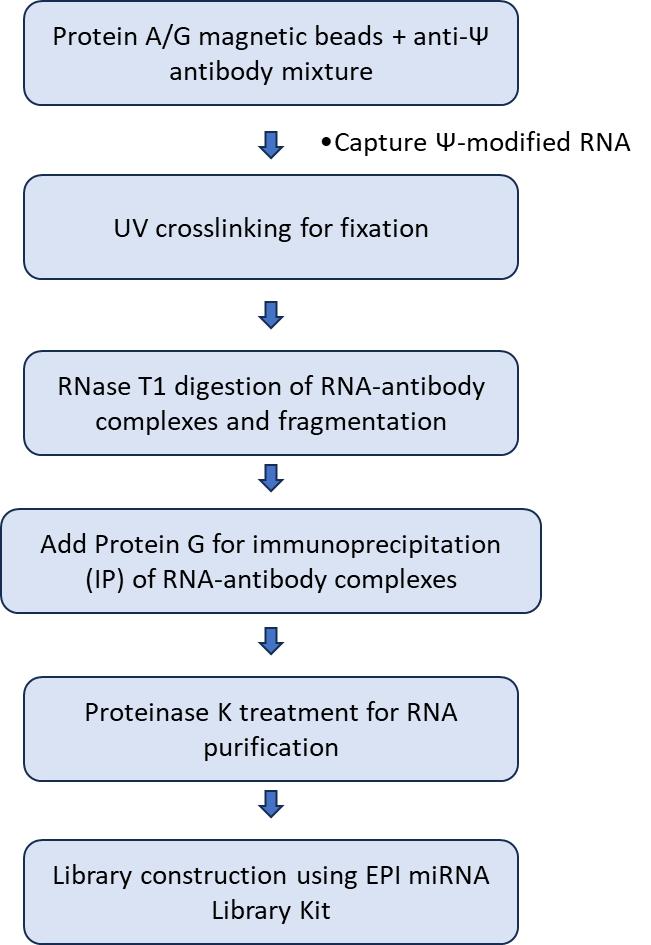 Figure 1. PA-Ψ-seq technical workflow
Figure 1. PA-Ψ-seq technical workflow
Service Workflow
Analysis Content
| Basic Analysis |
- Adapter trimming and quality control of raw sequencing reads.
- Alignment to the reference genome (Mapping).
- Identification of enriched regions (Peak Calling).
- Annotation of enriched regions (Peak Annotation).
- Analysis of mRNA modifications.
- Metagene plots, pie charts, and Venn diagrams for enriched regions.
- Motif analysis.
- Gene ontology (GO) and KEGG pathway analysis for peak-associated genes.
- Differential peak analysis (i.e., differential RNA modification in mRNA).
- GO and KEGG analysis for differential peak-associated genes.
|
| Advanced Analysis |
- Heatmap of differential RNA modifications.
- Correlation analysis (with RNA-Seg or multi-omics correlation analysis).
- Heatmap of correlation analysis.
- IGV peak visualization.
|
Service Advantages
- High Resolution: Achieves single-nucleotide resolution for precise mapping of Ψ modifications
- High Specificity: Antibody-based capture combined with UV crosslinking minimizes noise and enhances data accuracy
- Comprehensive Analysis: Extensive analysis from modification sites to functional pathways, providing a complete understanding of Ψ's role in gene regulation
Our Capabilities
- Precision in RNA Modification Detection: Our expertise lies in the precise identification of pseudouridine modifications at a single-nucleotide resolution, thereby enabling high-resolution insights into RNA modifications across the entire transcriptome.
- Customizable Workflow Design: From sample preparation to data analysis, our PA-Ψ-seq service is fully customizable to cater to the specific requirements of individual research projects, offering unparalleled flexibility in study design and experimental aims.
- Expertise in Complex Data Interpretation: Our bioinformatics team provides advanced, publication-ready data analysis encompassing peak annotation, motif discovery, and pathway enrichment analysis. This service aids researchers in elucidating the functional roles of RNA modifications.
- Advanced Crosslinking and Immunoprecipitation Techniques: Employing state-of-the-art photo-crosslinking and immunoprecipitation methods, our laboratory ensures the selective capture of Ψ-modified RNA with high specificity and minimal background interference.
- Scalable Solutions for High-Throughput Studies: Whether your research entails a few samples or a comprehensive transcriptomic analysis, we offer scalable solutions that ensure efficient data processing and high-quality results for both small and large-scale studies.
Applications
- Transcriptome-wide Ψ modification mapping
- Investigating the role of Ψ in cancer, neurological disorders, and other diseases
- Ψ modification correlation with RNA splicing and translation regulation
- Exploration of Ψ's role in immune response and antiviral defense mechanisms
Sample Requirements
Sample Types:
- Cells: A minimum of 6 × 10^7 cells per sample.
- Tissues: A minimum of 150 mg per sample.
- Total RNA: A minimum of 300 μg per sample, with a RIN value of ≥ 6.
Sample Species:
- Human and mouse samples are accepted. Other species require prior evaluation.
Deliverables
- High-quality raw sequencing data
- Comprehensive pseudouridine modification enrichment maps and differential analysis reports
- Functional enrichment analyses and visualizations (e.g., GO, KEGG)
- Detailed experimental and analytical reports
Our Features
CD Genomics has developed a cutting-edge RNA modification sequencing platform tailored for pseudouridine detection. Our expertise in PA-Ψ-seq ensures that your research goals are met with precision and reliability.
CD Genomics is a trusted partner for scientists worldwide, offering unparalleled technical support and high-quality sequencing services backed by years of experience in RNA modification research.
Why Choose Us?
CD Genomics is a company that provides professional and comprehensive MeDIP sequencing services. We have years of experience to meet your specific project needs in using epigenomics research to add value to your research projects. CD Genomics can provide you with personalized solutions to help you thrive every step of the way around your interest in your workflow. If you would like to know more about this service, please feel free to contact us.
! For research purposes only, not intended for clinical diagnosis,
treatment, or individual health assessments.

 Figure 1. PA-Ψ-seq technical workflow
Figure 1. PA-Ψ-seq technical workflow 