In 5-methylcytosine (m5C) RNA modifications, a methyl group attached to the fifth carbon of the cytosine ring in RNA molecules. m5C is likewise a widespread RNA modification detected in mRNA and non-coding RNA, which affect the regulate stress response, control translation efficiency, ,stability of the mRNA molecule and so on. With recent advances in sequencing technology and experimental methods, methylation modifications of RNA can be qualitatively and quantitatively analyzed at the single-nucleoside level, providing novel insights into its biological functions. CD Genomics adopts detection methods for m5C RNA modifications include RNA bisulfite sequencing (RNA-BisSeq) and m5C meRIP.
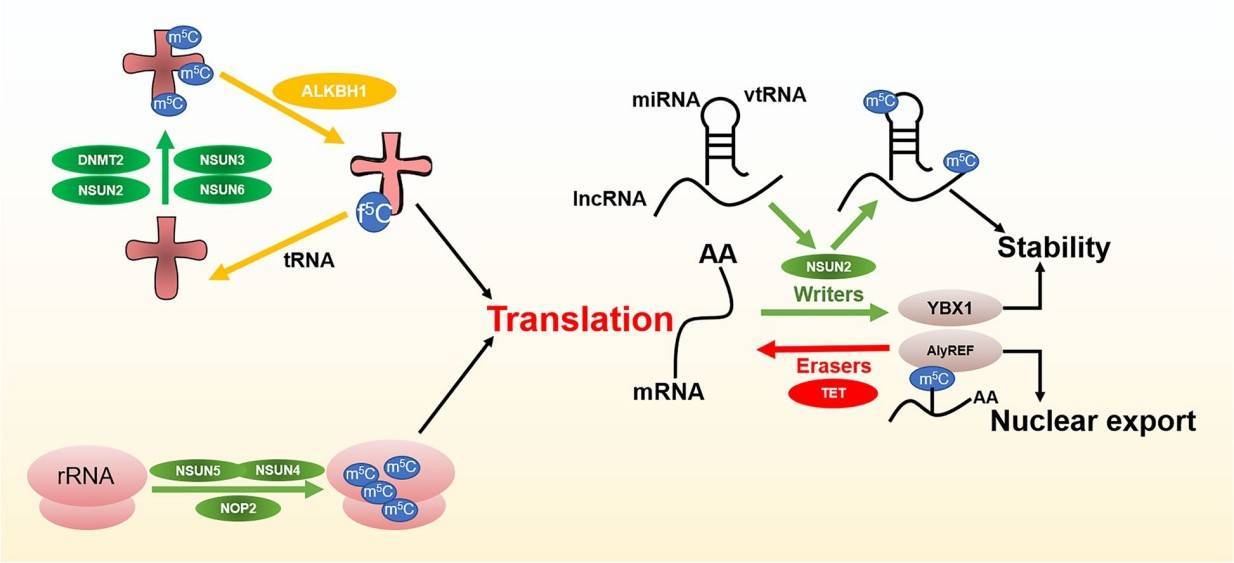 Figure 1. m5C modifcation in varied of RNA (Song,2022)
Figure 1. m5C modifcation in varied of RNA (Song,2022)
Specific m5C RNA-seq Approaches We Offer
Methods
- m5C meRIP
- RNA fragments were randomly interrupted
- The RNA m5C methylated specific antibody was co incubated with RNA fragments
- Enriched m5C methylated modified fragments were sequenced
- m5C RNA-BisSeq
- RNA fragments were randomly interrupted
- Bisulfite conversion of RNA
- Purification of bisulfite converted RNA
- Desulfonation and reverse transcription amplification
- Sequencing
Our m5C RNA-seq are available for a wide range of sequencing applications:
- Complete transcriptome sequencing of m5C
- Sequencing of m5c cyclic RNA
- m5C LnRNA sequencing
- m5C mRNA sequencing
Our bioinformatics analysis services include, but are not limited to:
- Sequencing data quality control
- Comparison with reference genome
- Identified methylated sites
- Length distribution of methylated sites and motif analysis results
- Differential methylated sites
- GO and Pathway analysis results of differential methylated sites
Service Advantages
- Specific enrichment and detection of RNA fragments methylated by m5C
- RNA m5C methylated detection in the whole transcriptome
- Targeted detection of m5C methylated of different types of RNA molecules
- RNA-BisSeq can identify the m5C modified site with single base resolution
- m5C meRIP can specifically enrich m5C modified RNA fragments
Our Capabilities
- Wide detection range: Whole-transcriptome wide RNA m5C methylated profiling.
- Flexible sample requirements: Cells, tissues or RNA are acceptable
- Specialized bioinformatics analysis: Strong bioinformatic team, professional m5C methylated data analysis.
Sample Requirements
The number of cells shall not be less than 2×107.
The quantity of the tissue samples should be between 100 mg-1 g.
The quantity of RNA sample should be no less than 30 μg,RNA purity: OD260/280 = 1.6-2.3; RNA quality: 28S:18S ≥ 1.5, RIN ≥ 7.
Service Process
Deliverables
- Related experimental results raw data
- Experimental report
- Data analysis
- Image and result analysis
- Bioinformatics analysis results
- Details in m5C -Seq for your writing
Our Features
- CD Genomics will complete your project on time and efficiently. We have professional after-sales service.
- CD Genomics works with scientists from many biotechnology companies. We have extensive knowledge and experience to provide quality assurance services.
Why Choose Us?
CD Genomics is a company that provides professional and comprehensive m5C -Seq services. We have years of experience to meet your specific project needs in using epigenomics research to add value to your research projects. CD Genomics can provide you with personalized solutions to help you thrive every step of the way around your interest in your workflow. If you would like to know more about this service, please feel free to contact us.
References
- Guo, Gangqiang, et al. Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance. Molecular Therapy-Nucleic Acids 26 (2021): 575-593.
- Song, Hang, et al. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. Biomarker Research 10.1 (2022): 1-15.
! For research purposes only, not intended for clinical diagnosis,
treatment, or individual health assessments.

 Figure 1. m5C modifcation in varied of RNA (Song,2022)
Figure 1. m5C modifcation in varied of RNA (Song,2022)