We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

TCR sequencing is a high-throughput sequencing technique, which is used to analyze the diversity, clonal distribution of TCR and its role in immune response. TCR is an important molecule on the surface of T cells, which is responsible for recognizing antigen peptide-major histocompatibility complex (MHC) complex, thus mediating immune response. TCR sequencing technology reveals the diversity and functional status of T cells by detecting the sequences of variable region (V region) and constant region (J region) of TCR gene.
At present, TCR sequencing has two mainstream technologies, namely multiplex PCR and 5'RACE. Multiplex PCR can be carried out for genomic DNA or RNA that has been transformed into double-stranded complementary DNA (cDNA). Primer pairs targeting J and V alleles are used to amplify CDR3 region of TCR transcript. After several rounds of PCR amplification, linkers are connected to both ends of the transcript. 5'RACE is mainly used to obtain the complete sequence of TCR, and its amplification preference is relatively low. In order to solve the problems of quantitative accuracy caused by repeated introduction of PCR and false positive TCR caused by PCR/sequencing amplification, some technologies will introduce UID digital tag error correction technology to accurately restore the abundance of TCR clones and remove false positive TCR.
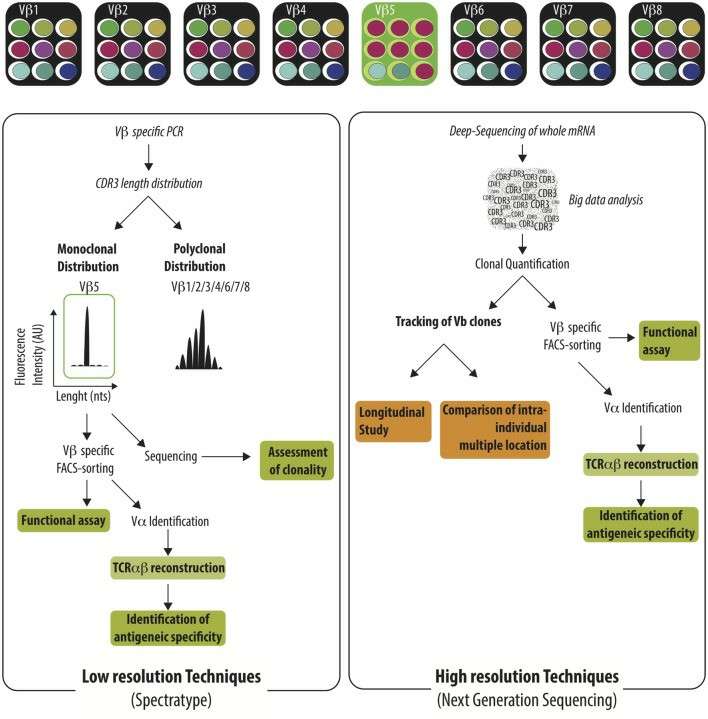 Characterization of the TCR vβ repertoire by low resolution and high resolution technique (Degauque et al., 2016)
Characterization of the TCR vβ repertoire by low resolution and high resolution technique (Degauque et al., 2016)
TCR sequencing is a high-throughput sequencing technique used to analyze the diversity and function of TCR. Its principle is to extract RNA or DNA from T cells and sequence TCR gene by high-throughput sequencing technique to analyze the diversity and clonality of T cell receptor. The specific process is as follows.
Sample collection and cell separation: Firstly, it is needed to collect samples containing T cells, such as peripheral blood and tissue biopsy samples. Then, T cells are separated from the sample by density gradient centrifugation, magnetic bead sorting, flow cytometry and other methods, which can enrich specific types of T cells (such as CD4+ or CD8+ T cells) and remove unnecessary cell types.
Nucleic acid extraction: Total RNA or DNA is extracted from T cells. RNA sequencing (RNA-seq) is mainly used to analyze transcripts of TCR α and β chains, while DNA sequencing directly amplifies TCR genes. In order to improve the efficiency of sequencing, PCR amplification technology, such as 5' RACE or multiplex PCR, is usually used to ensure the specificity and efficiency of amplification.
PCR amplification: TCR gene consists of multiple variable region (V), diverse region (D), junction region (J) and constant region (C) gene fragments. Primers were designed according to the conserved sequence of TCR gene, and the V-D-J region of TCR was specifically amplified by PCR. Because TCR genes of different T cells are different in V, D and J regions, a large number of DNA fragments with different lengths will be obtained after PCR amplification, which represent different TCR clones.
High-throughput sequencing: The DNA fragments amplified by PCR were subjected to high-throughput sequencing. At present, the commonly used sequencing platforms are Illumina and PacBio. These platforms can simultaneously sequence a large number of DNA fragments and generate massive sequencing data.
Data analysis: The sequencing data were analyzed by bioinformatics methods. Firstly, the original sequencing data are quality controlled and filtered to remove low-quality sequences. Then the processed sequences are compared with the TCR gene database to determine the V, D and J gene fragments corresponding to each sequence, so as to analyze the diversity of TCR, the types of clones and the frequency of different clones. The amino acid composition and structural characteristics of TCR sequence can be further analyzed to understand the function and immune response mechanism of T cells.
Through TCR sequencing, we can deeply understand the immune state of T cells, and provide important theoretical basis and technical support for the study of immune-related diseases, tumor immunotherapy, vaccine development and so on.
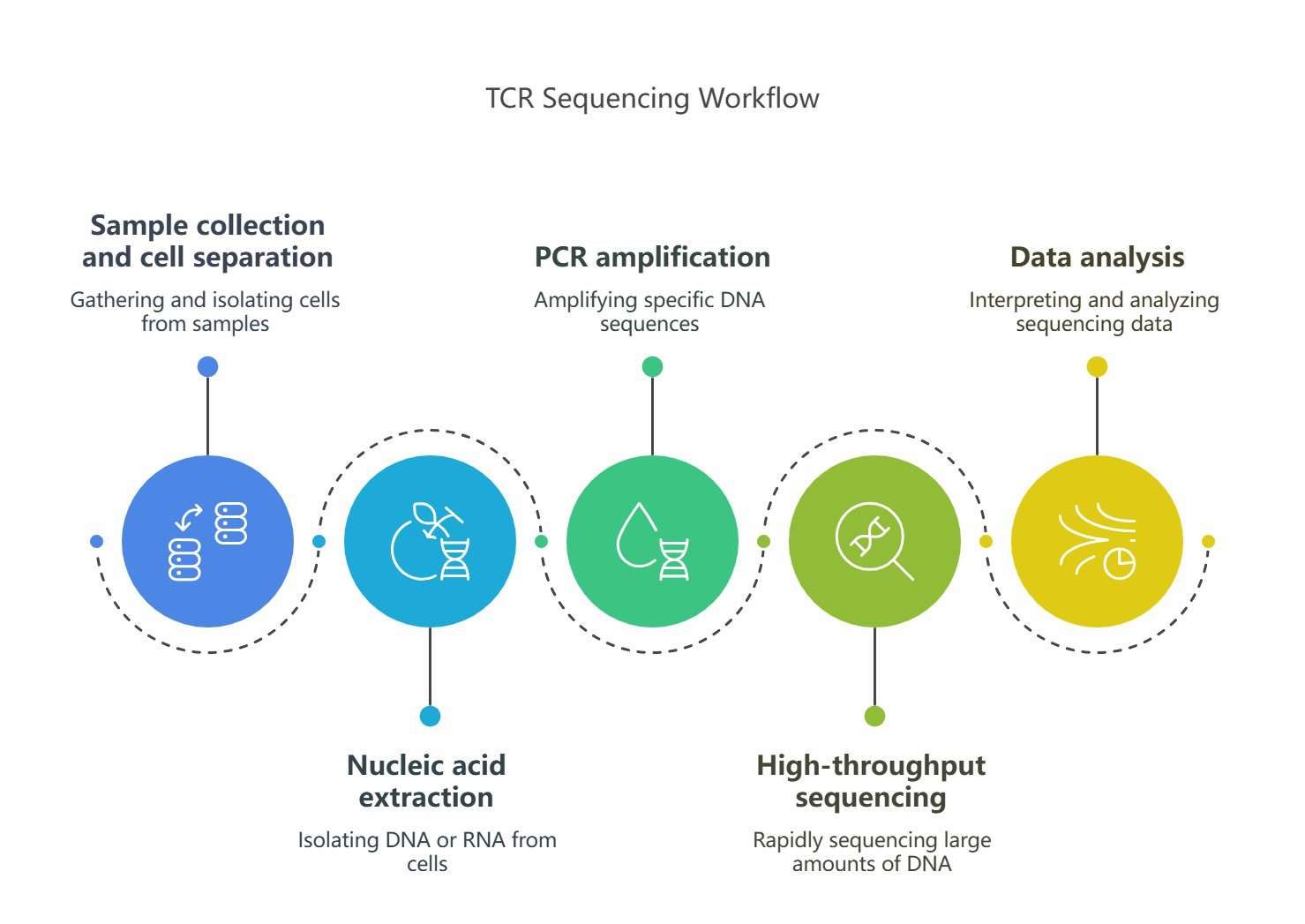 TCR sequencing workflow
TCR sequencing workflow
The analysis of TCR sequencing results is a complex but important work, and its main applications include tumor immunotherapy, immune microenvironment research, disease diagnosis and treatment effect evaluation, etc. The following are its main analysis steps and contents.
TCR gene identification and annotation: The pretreated sequenced reads are compared to the reference sequence of human TCR gene, and the common tools such as IMGT/HighV-QUEST are used to determine which TCR gene fragment (such as V, D and J gene fragments) each read comes from. Detailed annotation information of TCR gene fragments, including gene name, function, sequence characteristics, etc., is obtained through related databases (such as IMGT database), which provides a basis for subsequent analysis.
Clonal analysis: the clonal type of TCR is determined by its unique combination of V, D and J gene fragments and the amino acid sequence of CDR3 region. By analyzing the distribution of different clones in sequencing data, we can understand the diversity and specificity of T cell population. The frequency of each clone in sequencing data was counted to evaluate its relative abundance in T cell population. Clones with high frequency may represent specific T cell amplification for specific antigens.
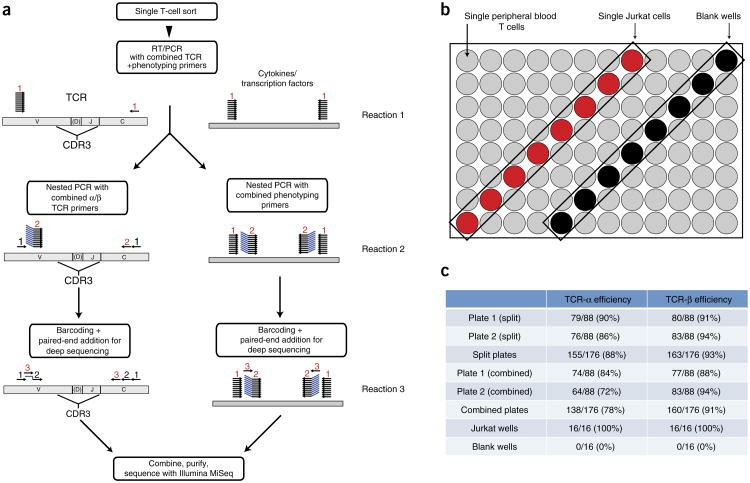 Strategy for single-cell TCR sequencing and phenotyping, and determination of TCR-sequencing efficiency (Han et al., 2014)
Strategy for single-cell TCR sequencing and phenotyping, and determination of TCR-sequencing efficiency (Han et al., 2014)
Difference analysis: If there are multiple samples (such as disease group and healthy control group), the difference analysis between groups can be carried out to find out TCR clones or gene fragments that are significantly differentially expressed in different groups. This is helpful to find specific T cell subsets related to diseases. In order to further reveal its biological significance, the differentially expressed TCR clones were analyzed by functional enrichment, such as whether they were associated with specific immune pathways or disease-related gene sets.
Take the Next Step: Explore Related Services
Learn More
TCR sequencing is a high-throughput sequencing technique used to analyze the diversity and function of TCR. It has a wide range of applications, including immunology research, disease diagnosis, treatment monitoring and immunotherapy development.
Autoimmune diseases: TCR sequencing is helpful to analyze the characteristics of abnormally activated T cell subsets and their receptors in autoimmune diseases, clarify the clonal types and distribution of autoreactive T cells, thus deeply understanding the pathogenesis of diseases and providing new targets and strategies for the diagnosis and treatment of diseases.
Tumor immune monitoring: TCR sequencing is used to dynamically monitor the changes of T cells in patients during tumor treatment, and to evaluate the efficacy of immunotherapy and the risk of tumor recurrence. For example, observing the cloning and amplification of tumor-specific T cells and the changes of TCR diversity after treatment can adjust the treatment plan in time.
Efficacy evaluation of immunosuppressants: TCR sequencing can analyze the influence of immunosuppressants on the recipient's T cell receptor pool, evaluate the inhibitory effect of drugs on T cell immune function, guide the individualized use of immunosuppressants, reduce the side effects of drugs, and ensure the immune tolerance of transplanted organs.
Discovery of immune drug targets: a comprehensive analysis of the interaction between TCR and antigen is helpful to discover new immune regulatory targets and provide a basis for developing new immunotherapy drugs. For example, develop small molecular inhibitors or biological agents for specific TCR subtypes or key molecules in their signaling pathways to regulate abnormal immune responses.
Drug efficacy prediction: TCR sequencing is used to analyze the characteristics of T cell receptors of patients before treatment, predict patients' response to immunotherapy drugs, and realize precise medical treatment. For example, some patients with high TCR diversity or specific TCR clones may have a better therapeutic effect on immune checkpoint inhibitors, and TCR sequencing can help screen out such dominant groups and improve the effective rate of treatment.
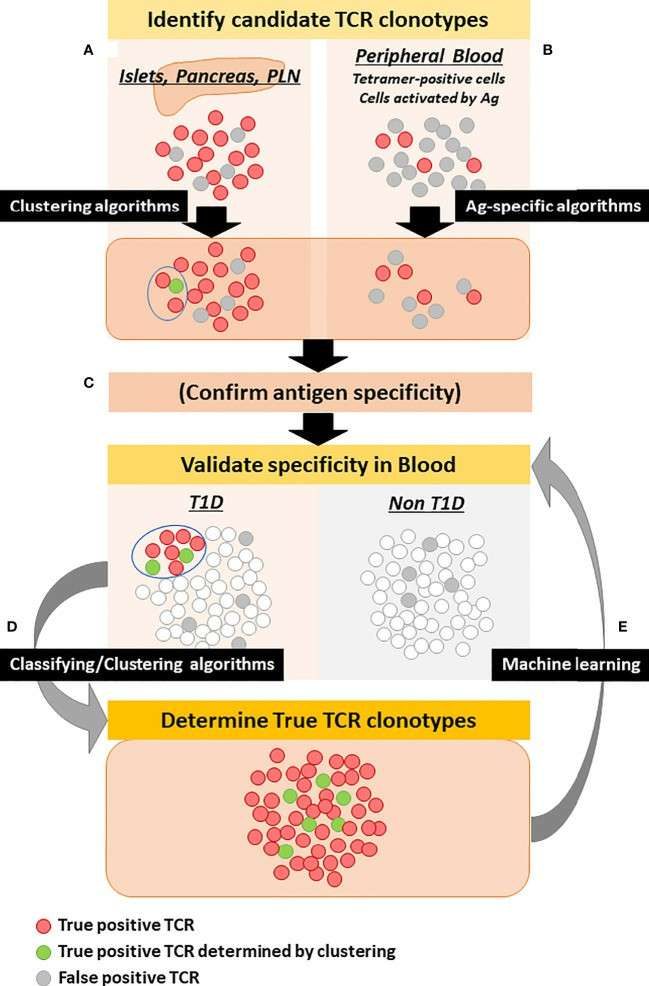 Strategy to determine disease-specific TCR clonotypes (Nakayama et al., 2021)
Strategy to determine disease-specific TCR clonotypes (Nakayama et al., 2021)
Identification of rare and low-frequency clones: It is highly sensitive and can detect rare T cell clones which account for a very low proportion in the whole T cell population. Even those specific T cells, which account for only a small proportion in the body, may be accurately identified and analyzed, which is very important for studying some specific immune responses against rare pathogens or tumor cells.
Tracking the dynamic changes of T cells: The dynamic changes of T cell population can be clearly observed by TCR sequencing of samples at different time points or in different physiological and pathological states. For example, in the process of infection, we can track how T cells clone, amplify and differentiate against pathogens; In the process of tumor treatment, the immune response of T cells to tumor antigen can be monitored, so as to evaluate the therapeutic effect and predict the recurrence of the disease.
Helping disease diagnosis and monitoring: In many diseases, the clonality and diversity of T cells will undergo characteristic changes. TCR sequencing can help doctors diagnose and monitor many diseases, such as assisting in the diagnosis of hematological malignancies, such as lymphoma and leukemia, by detecting the clonal proliferation of TCR. It can also be used to monitor the immune state after organ transplantation and predict the occurrence of rejection.
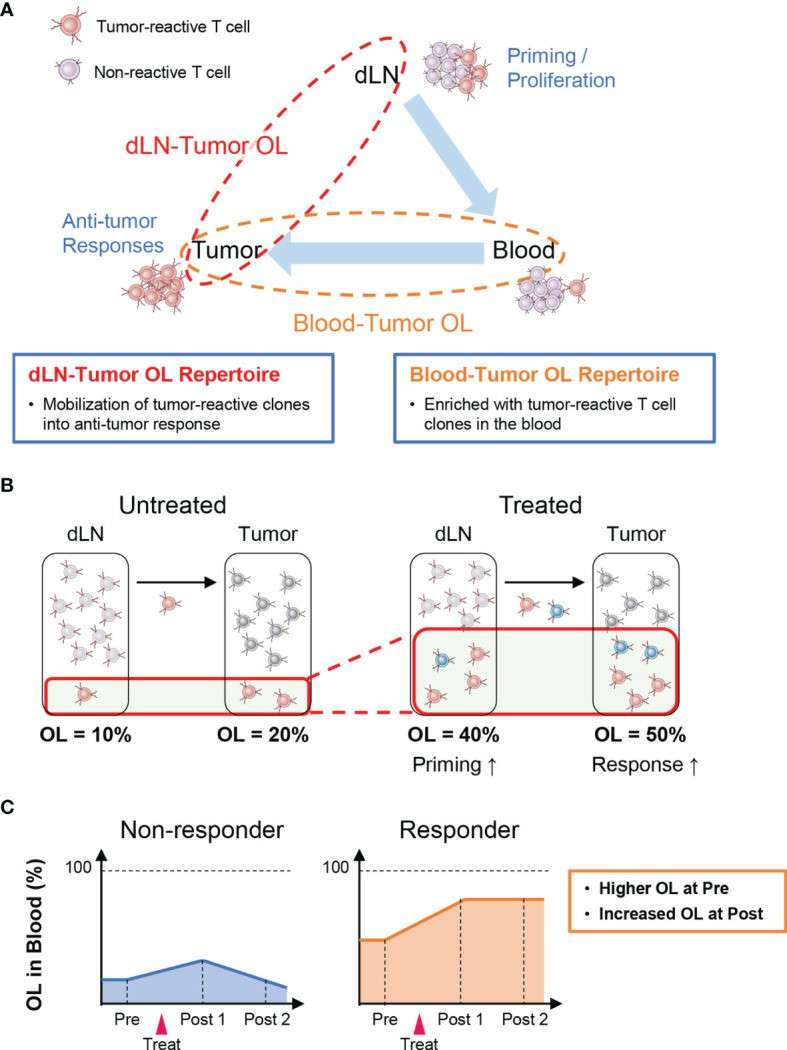 Outline of inter-organ clone tracking analysis (Aoki et al., 2022)
Outline of inter-organ clone tracking analysis (Aoki et al., 2022)
TCR sequencing has broad future prospects in technical development, clinical application and scientific research. The following are some specific prospects.
A. Technical development
a) In-depth analysis of single cell level: At present, single cell TCR sequencing has made some progress, and this technology will be more mature and popular in the future. It can not only provide TCR sequence information of a single T cell, but also combine with single cell transcriptomics, protein genomics and other technologies to comprehensively analyze the function, state and interaction of T cells with other cells at the single cell level, and deeply understand the dynamic changes and mechanism of T cells in immune response.
b) Multiomics data integration: TCR sequencing will be more widely and deeply integrated with genomics, transcriptomics, protein genomics, metabonomics and other technologies. By comprehensively analyzing multi-dimensional data, a more comprehensive immune map is constructed, which systematically reveals the regulatory network and molecular mechanism of the immune system in health and disease States, and provides a more comprehensive basis for disease diagnosis, treatment and drug research and development.
B. Clinical application
a) Guide personalized immunotherapy: Based on the unique TCR spectrum of patients, more targeted immunotherapy strategies can be developed. For example, design personalized T cell receptor engineered T cell (TCR-T) therapy, select TCR clones with high affinity and specificity for gene editing, and enhance the ability of T cells to recognize and kill tumor cells or pathogens. In addition, TCR sequencing can also help to screen patients who are suitable for immune checkpoint inhibitor treatment, and predict patients' response to treatment, realize precise medical treatment, improve treatment effect and reduce adverse reactions.
b) Vaccine development and evaluation: In vaccine development, TCR sequencing can be used to evaluate the immune response of the body after vaccination. By monitoring the dynamic changes of T cell clones, we can understand the formation mechanism of immune memory induced by vaccines, optimize vaccine design and vaccination scheme, and improve the immunogenicity and protective effect of vaccines. At the same time, TCR sequencing can also be used as an important means to evaluate the safety and effectiveness of new vaccines, such as mRNA vaccines, and provide strong support for the clinical application of vaccines.
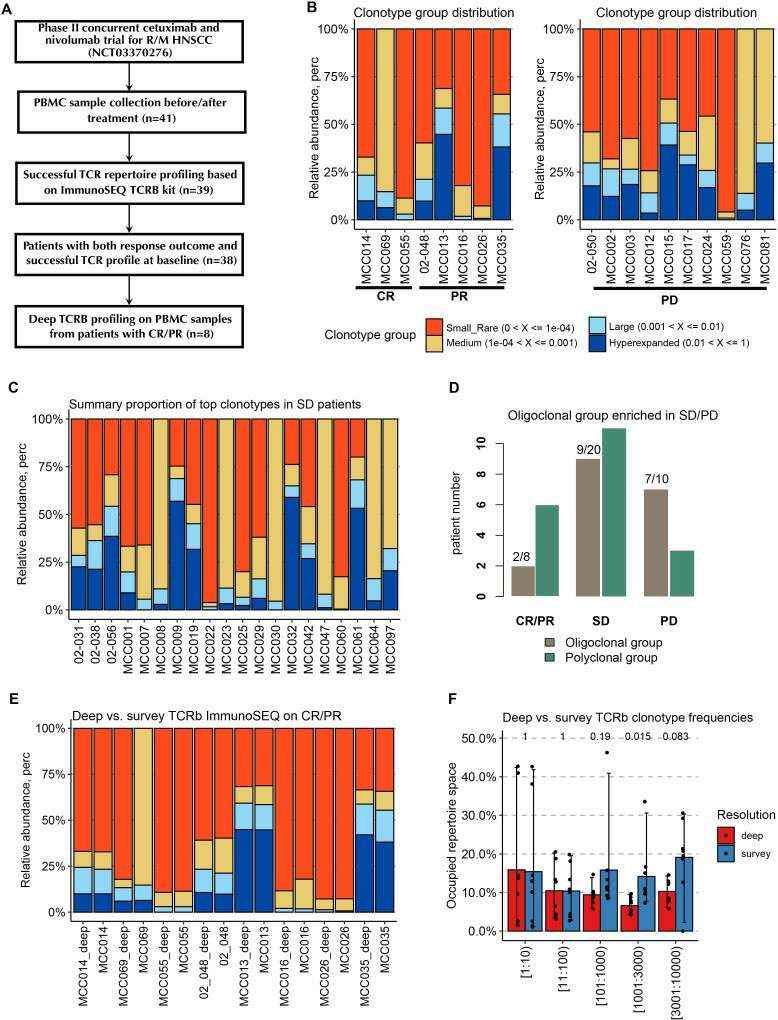 Patients with CR/PR have a less oligoclonal peripheral TCR expansion prior to the combination therapy (Wang et al., 2022)
Patients with CR/PR have a less oligoclonal peripheral TCR expansion prior to the combination therapy (Wang et al., 2022)
A. Scientific research and exploration
a) Study on immune development and evolution: TCR sequencing will provide a deeper insight into the development, differentiation and maturation of T cells. By analyzing the TCR spectra of T cells at different developmental stages, the dynamic changes and regulation mechanism of TCR gene rearrangement are revealed, which is helpful to understand the ontogeny and evolution of immune system. In addition, comparing the TCR sequence and diversity of different species can also explore the conservatism and adaptability of the immune system in the evolution process, and provide a new perspective for the basic research of immunology.
b) Study on the mechanism of immune-related diseases: For various immune-related diseases, such as infectious diseases, allergic diseases and autoimmune diseases, TCR sequencing can help to deeply analyze the immunopathological mechanism of disease occurrence and development. By comparing the TCR spectra of patients and healthy controls, we can find the specific T cell clones and immune characteristics related to diseases, reveal the abnormal activation, proliferation and differentiation process of immune cells in diseases, provide key clues for the pathogenesis research of diseases, and promote the basic research of related diseases and the development of translational medicine.
TCR sequencing, as a key technical means to accurately analyze the function and diversity of T cells in immune system, has shown great value in immunology research and clinical application. In terms of methods, by highly sensitive amplification of TCR gene and high-throughput sequencing, the variable region sequence of T cell receptor can be accurately captured, and the complexity and diversity of T cell bank can be fully revealed.
In the future, with the continuous development of sequencing technology and the improvement of analytical methods, TCR sequencing will play a more important role in immunology and medicine, providing new ideas and methods for overcoming more complex diseases.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy