We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

T cells are part of the adaptive immune system, and their responses are mediated by a unique T cell receptor (TCR), which can recognize specific antigens from various biological environments. Therefore, the analysis of T cell bank can better understand immune response and cancer and other diseases. The second generation sequencing technology has greatly realized the Qualcomm analysis of TCR library. Based on the rich experience in this field in the past ten years, the author provides an overview of TCR sequencing, from the initial library preparation steps to sequencing and analysis methods, and finally to functional verification technology. As for data analysis, the author introduces the important indexes of TCR library in detail, and puts forward several calculation tools to predict antigen specificity.
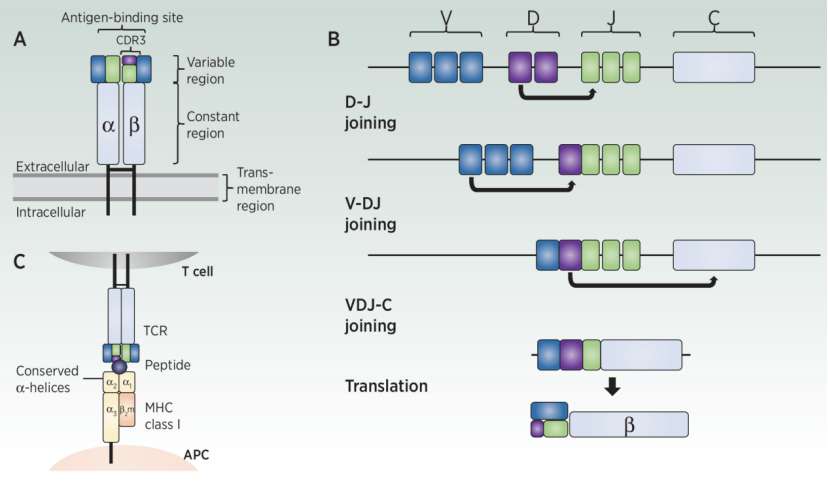 Schematic representation of TCR, MHC, and V(D)J recombination (Meredith et al., 2023)
Schematic representation of TCR, MHC, and V(D)J recombination (Meredith et al., 2023)
TCR is a highly diverse heterodimer surface receptor, which enables T cells to provide protection against various pathogens. The functional TCR consists of two paired protein chains, including α chain and β chain, or γ chain and δ chain.
Most T cell TCRαβ chains can recognize antigens presented on MHC proteins. Only 1%-5% of T cells are not restricted by MHC and participate in innate immune response. The exact ligand for T cell binding is unknown. For these reasons, α β TCR is usually the focus of TCR sequencing and subsequent clinical application.
The variable domain of each TCR chain has three complementarity determining regions (CDR): CDR1, CDR2 and CDR3. CDR1 and CDR2, encoded by V gene fragment, promote the interaction between TCR and MHC mainly through conservative a- helix contact with MHC. CDR3 is encoded between V and J or D and J gene fragments, resulting in high variability. This region is responsible for binding MHC-presented antigenic peptides. Because of its direct interaction with antigen and inherent high variability, CDR3 region provides rich diversity for the specificity of TCR, so it is a common target region for TCR sequencing.
TCR specificity is the basis of understanding the biology and dynamics of T cells by TCR sequencing, as well as studying T cells in disease background and providing information for the development of treatment methods. Although the same TCR may be more inclined to recognize the same antigen, cross reactivity has also been observed, in which a single TCR has the ability to recognize multiple pMHC pairs. Because the cross-reactivity of TCR may complicate the characterization and monitoring of T cell population, it can not be ignored when analyzing TCR sequencing and verifying antigen specificity.
Multiplex PCR and 5'RACE database construction
In the preparation of T cell bank, there are two commonly used targeted amplification methods, namely multiplex PCR and 5'RACE, which usually mainly amplify CDR3 region of TCR.
Multiplex PCR primer library: Multiplex polymerase chain reaction is widely used, which involves using J gene fragment allele primer or TCR constant region primer to combine with the mixture of known V gene fragment alleles to amplify gDNA or RNA in CDR3 region. Essentially, multiplex PCR is limited by available primers. Because of this, the new V allele cannot be accurately expressed. In addition, primer bias in multiplex PCR will lead to uneven allele amplification and inaccurate relative TCR sequence frequency. By designing and modifying the double bar code and molecular barcode, it is helpful to remove the products introduced in the PCR amplification process and support accurate downstream sequence analysis.
5'RACE Single Primer Database Building: 5'RACE uses RNA and only uses a pair of primers to target the constant region of TCR chain and the 5' end of mRNA, which eliminates the bias caused by multiplex PCR and allows all TCR in a given sample to be captured.
Bulk sequencing and single cell sequencing
As for sequencing methods, bulk sequencing or single cell sequencing can be used to analyze T cell banks.
Bulk sequencing: It sequences the aggregation of unpaired TCR chains in a sample, and is usually used to analyze the diversity of TCR on a large scale and compare people in patient queues. Historically, batch analysis methods have mainly focused on the sequencing of TCRβ chain (or δ, γ δ in T cells), because there are additional D gene fragments and greater combinatorial diversity compared with α chain. In addition, T cells participate in "allele rejection", in which case only one functional β chain exists in T cells, and multiple α chains may also be expressed. However, one of the main limitations of sequencing only β -chain is that it can not provide information about β -chain pairing or the biological function of T cells after sequencing.
Single cell sequencing: Single cell sequencing focuses on a single immune cell. The progress of single cell separation technology, such as fluorescence activated cell sorting and microfluidic platform, laid the foundation for the success of single cell TCR sequencing. Although bulk sequencing provides valuable information on TCR diversity, it lacks the resolution of single cell analysis of all TCRαβ chain pairing.
Index characteristics of TCR library
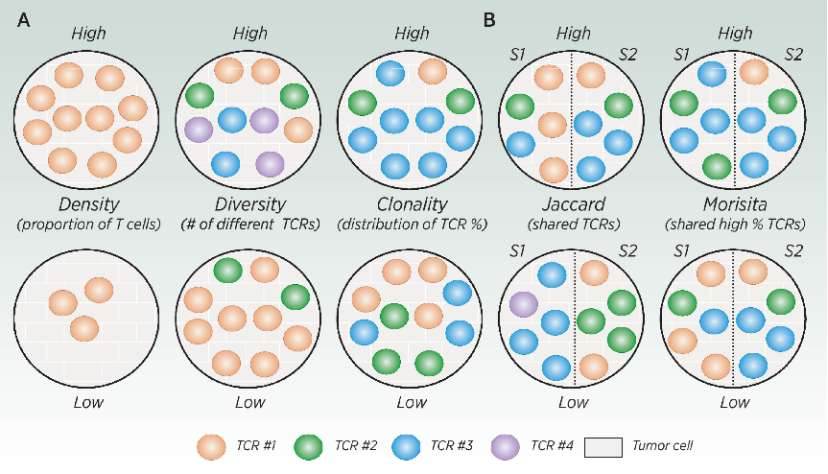 T cell bank indicators (Meredith et al., 2023)
T cell bank indicators (Meredith et al., 2023)
T cell density, diversity and cloning index show the characteristics of T cell bank. Compared with the high index and the low index, the color represents different types of T cell clones. Density refers to the proportion of T cells in a given area, diversity refers to the number of unique clones in the sample, and clonality refers to the amplification of clones in the sample.
In addition to reporting the indicators of individual sample attributes, there are also some indicators that are helpful to compare T cell banks between different samples. Two commonly used similarity indexes are Jaccard index and Morisita overlap index (MOI). Related indexes such as Jaccard index and Sorensen index are only based on whether there is a specific TCR sequence in multiple samples. These methods are useful in comparison, but they cannot compare the relative frequencies of specific TCR sequences between samples. In contrast, MOI explained the relative frequency and the existence of specific TCR sequences, providing a more comprehensive comparison of T cell libraries from different samples.
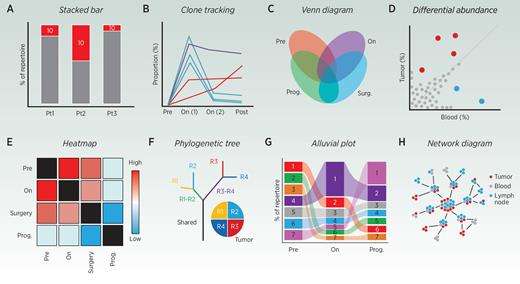 Common representations of TCR-related data (Meredith et al., 2023)
Common representations of TCR-related data (Meredith et al., 2023)
Clonal type (a), clonal tracking map (b), venn diagram (c), differential abundance map (d), thermal map (e), phylogenetic tree (f), alluvial map (g) and network map (h); Are used to represent TCR data. These charts provide information about the frequency and evolution of T cell clones in different patients, different tissue types, different diseases or treatment conditions.
TCR and antigen database
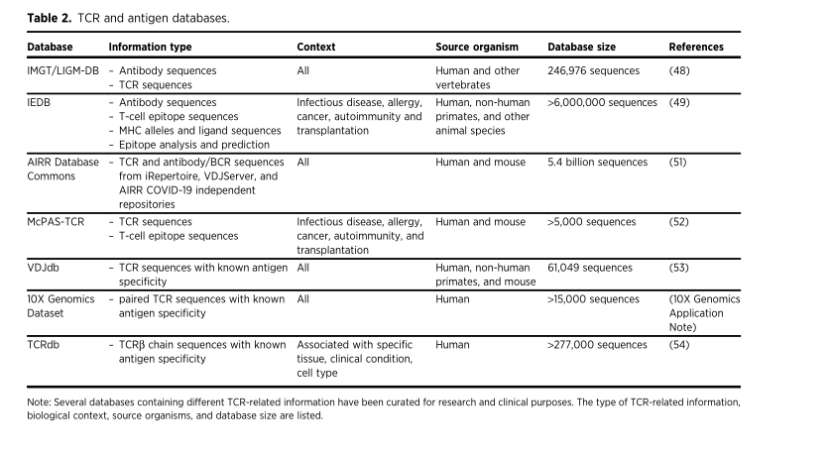 TCR and antigen databases (Meredith et al., 2023)
TCR and antigen databases (Meredith et al., 2023)
These databases are the basis of many computing tools, which are used to analyze TCR libraries and important aspects of immune response.
Computational tool for predicting antigen specificity
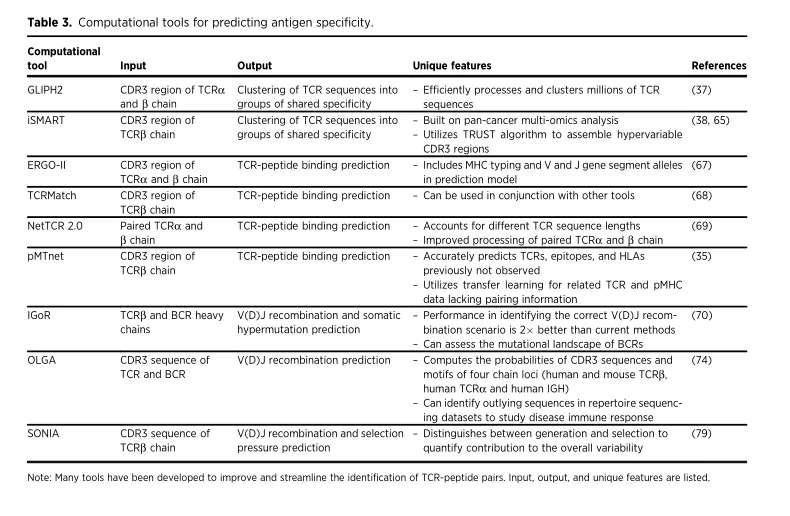 Computational tool for predicting antigen specificity (Meredith et al., 2023)
Computational tool for predicting antigen specificity (Meredith et al., 2023)
TCR sequencing not only describes the function of T cell bank through specific indicators, but also provides information for determining the specificity of T cell antigen. This information can help us better understand the reaction of T cells to specific antigens in individuals, so as to better understand and treat some diseases. At the same time, this information also provides a basis for designing customized immunotherapy methods. In the field of cancer immunology, people pay attention to identifying and targeting new antigens and peptides produced by somatic mutations unique to cancer cells. With the recent progress of HTS technology, many teams, including the author himself, have developed tools based on machine learning and algorithm to clarify antigen specificity.
Take the Next Step: Explore Related Services
Expression of tumor characteristics
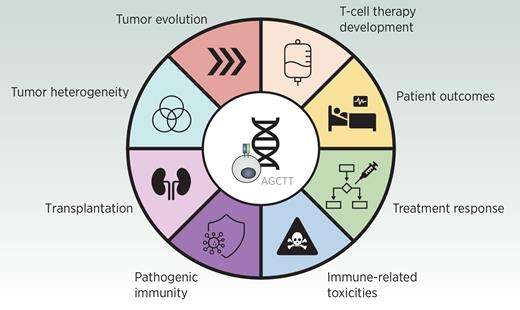 Applications of TCR sequencing and T-cell repertoire analysis (Meredith et al., 2023)
Applications of TCR sequencing and T-cell repertoire analysis (Meredith et al., 2023)
TCR sequencing promotes the expression of immune tumor heterogeneity, which is related to the genomic heterogeneity of tumor cells in the process of tumor occurrence. The authors sequenced TCR in non-small cell lung cancer (NSCLC) and found that there were significant differences in T cell density and clonality among different tumor regions, and there was a correlation between genomic and immune heterogeneity in tumor, indicating that TCR intratumor heterogeneity may be driven by specific antigens in different tumor regions.
Recently, some researcher also described the heterogeneity of TCR library in NSCLC, and provided more evidence that the heterogeneity in TCR tumor reflected the mutation in different tumor regions. Significant intra-tumor heterogeneity of TCR was also observed in non-small cell lung cancer metastasis and other cancers such as melanoma and renal cell carcinoma.
In addition to characterizing intra-tumor heterogeneity, TCR sequencing can also be used to observe inter-tumor heterogeneity. Specifically, the author's team used TCR sequencing to observe heterogeneous T cell profiles of synchronous metastasis of lung cancer and melanoma, which correspond to heterogeneous mutation and methylation.
Through these studies, TCR sequencing of multiple synchronous tumors can evaluate the differential growth of tumors before treatment. In the same patient, in addition to the focus-specific treatment response, the transfer between targeted therapy and immune checkpoint blockade was in the same patient.
Based on recent observations, cancer-related TCR has similar biochemical characteristics to TCR in normal tissues. Beshnova and colleagues developed a machine learning method called DeepCAT (Deep CNN Model of Cancer-related TCR) to predict whether there is cancer-related TCR in patients' peripheral blood. DeepCAT shows high prediction accuracy in distinguishing cancer patients from healthy individuals, and AUC of several early cancers (including breast cancer, ovarian cancer and melanoma) is higher than 0.95. Overall, DeepCAT represents a potential noninvasive method for early cancer detection.
Development and characteristics of immunotherapy
TCR sequencing is necessary for current T cell therapy. Adoptive T cell transfer therapy used to rely on autologous infusion of expanded tumor infiltrating T cells. However, due to the low frequency of new antigen-specific T cells in tumor infiltration group, many patients did not respond to this treatment. The progress of T cell therapy, including genetically modified T cells expressing new TCRs, has been developed to trigger tumor-specific tumor cell immune response.
TCR sequencing plays an important role in improving the efficacy of T cell therapy by identifying the best target antigen and tumor antigen-specific TCR, TCR. The isolated new antigen-specific T cells can be sequenced by whole or single cell TCR to identify the dominant TCR clone in tumor.
The analysis of TCR library also enables researchers to clarify the potential mechanism of immunotherapy and its influence on immune cells. Sipuleucel-T is an autologous cell therapy and a cancer vaccine, in which APCs are activated to fight against prostate acid phosphatase antigen, which is widely expressed in prostate cance. Researchers used TCR sequencing to characterize the mechanism of sipuleucel-T. They found that treatment promoted the recruitment of T cells into prostate, rather than the expansion of T cell clones, because the diversity of circulating T cells decreased, and the diversity of tumor infiltration also reduced the increase of T cells.
Predict the prognosis, treatment response and toxicity of patients
In addition to providing information for the development of immunotherapy, TCR sequencing and derived TCR library indicators have practical value in predicting the prognosis of patients. TCR diversity represents a prognostic biomarker for melanoma patients. Among melanoma patients, patients with higher uniformity and richness of T cells in peripheral blood and lymph node metastasis have longer progression-free survival and overall survival.
The increase of TCR pool diversity of tumor infiltrating T cells has been found to have an impact on the prognosis of many cancers, including breast cancer, melanoma, lung cancer and kidney cancer. In a study on high-grade serous ovarian cancer, high T cell clonality combined with genomic instability features (including homologous recombination defects and copy number variation) or T cell infiltration into the tumor led to high prognostic value. In addition, patients whose expanded T cells have higher affinity for new antigens from high mutation allele frequency are more likely to show better prognosis and response to immunotherapy in lung cancer and melanoma.
Although cancer immunotherapy promotes significant anti-tumor immune response, they may also cause toxic reactions related to inflammation. In addition to being a biomarker for prognosis and predicting patients' prognosis and immunotherapy response, TCR sequencing can also provide information about potential immunotherapy-related toxic reactions. Clonal detection is related to immune-related adverse events (irAE) in prostate cancer patients receiving anti-CTLA-4 therapy. The amplification of more than 55 CD8+T cell clones in peripheral blood indicates the development of severe irAE. Another study correlated the density and diversity of CD4+ memory T cells in peripheral blood of patients with melanoma with the development of severe irAE with anti-PD-1 monotherapy and anti-PD-1 and anti-CTLA-4 combined therapy.
The usefulness of TCR sequencing is not limited to cancer applications. Because T cells and their TCR play an indispensable role in mammalian xenotransplantation by identifying and eliminating incompatible tissues, researchers used TCR sequencing to characterize the response of organ transplantation. Specifically, TCR library can provide information on rejection risk through T cell-mediated rejection and graft-versus-host disease, and has been evaluated in kidney, liver and hematopoietic stem cell transplantation.
In addition to transplantation, TCR sequencing has potential applications in biological characterization, disease stratification and autoimmune disease monitoring. For example, by analyzing the TCR gene pool, researchers have deeply understood the dynamics of spatial T cells in multiple sclerosis, determined the specific TCR clones of celiac disease, and defined clonal amplification and TCR chain pairing in type I diabetes.
T cells play an important role in the immune response to pathogens. Therefore, TCR sequencing is of great value for studying adaptive immune dynamics, monitoring the progress of infection and developing new immunotherapy for many different virus infections (including SARS-CoV-2). In other health conditions, such as ischemic heart disease and acute coronary syndrome, TCR pedigree analysis can be performed for risk stratification and diagnosis.
Generally speaking, this shows that TCR sequencing and TCR library can be used in many biological fields, and have a very broad application prospect in the biomedical field.
If you would like to learn more about the applications of TCR sequencing, you can refer to the following articles:
TCR sequencing is a tool to describe immune response, understand the nature of TCR library and help to identify TCR antigen pairs. Specific to cancer, TCR sequencing contributes to the basic characteristics of tumor heterogeneity and tumor evolution. From the clinical point of view, TCR sequencing is not only the basis of T cell immunotherapy, but also a tool to evaluate the efficacy of various immunotherapy. In addition, the TCR library index obtained from sequencing data analysis can be used as an important predictor of prognosis, anti-cancer treatment response and immune-related toxicity risk of many patients with different cancer types. The NGS method and TCR data analysis tools have been continuously improved, showing greater processing capacity and expanding research and clinical applications.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy