T Cell Receptor Sequencing: Applications in Immune Research
T cell receptor (TCR) is a molecule located on the surface of T cells, which plays a crucial role in specifically recognizing antigens and mediating immune responses. The diversity of the TCR repertoire, resulting from gene recombination and selective expression during T cell development, directly reflects the state of the immune system's response. TCR sequencing has emerged as a key technology for dissecting the function and specificity of T cells within the immune system.
The Principle of TCR Sequencing
Bulk TCR sequencing is commonly employed to analyze pooled T-cell populations. This approach allows for the investigation of TCR diversity and clonality and facilitates the comparison of TCR repertoires between different samples based on the frequency of single TCR chains. Current protocols for bulk TCR sequencing can utilize either genomic DNA (gDNA) or RNA extracted from fresh or formalin-fixed samples, each with its own set of advantages and disadvantages. Given that the variability, diversity, and joining (VDJ) complex constitutes a relatively small portion of the total gDNA, specific techniques are required for its analysis.
Structural basis of TCR gene: The structural basis of the TCR gene lies in its composition of either α and β chains or γ and δ chains. During T cell development, the genes undergo rearrangement of V, D, and J gene fragments in a random manner, generating a vast array of TCR sequences. This process endows T cells with the capacity to recognize a diverse range of antigens.
TCR gene amplification: To perform TCR gene amplification, T cell RNA is first extracted from samples such as blood or tissues and then reverse transcribed into cDNA. Two main methods are utilized for amplification: multiplex PCR, which employs multiple pairs of primers to target the V, D, and J regions, and 5'RACE. In 5'RACE, a universal linker is added to the 5' end of the cDNA, followed by amplification using the linker primer and TCR C region primer.
High-throughput sequencing: Subsequently, the amplified TCR gene products are subjected to high-throughput sequencing. Second-generation sequencing technologies like Illumina offer high throughput and relatively low cost but have shorter read lengths. In contrast, third-generation technologies such as PacBio can sequence the complete TCR variable region sequence with longer read lengths.
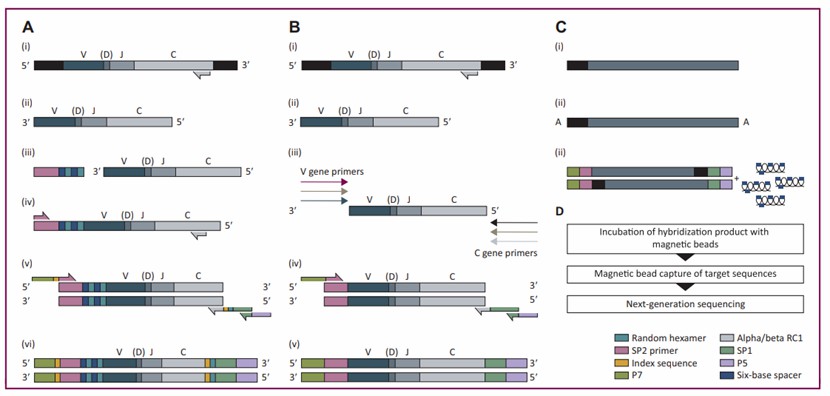 Schematic principle of TCR sequencing (Sanroman et al., 2023)
Schematic principle of TCR sequencing (Sanroman et al., 2023)
The Applications of TCR Sequencing
TCR sequencing has found extensive applications in various fields of immunology. In tumor immunology, it provides valuable insights into the characteristics of tumor-infiltrating T cells, aiding in the evaluation of treatment efficacy and the development of novel therapeutic strategies. In the research of infectious diseases, monitoring changes in the TCR repertoire enables the understanding of the dynamic immune response, guiding treatment decisions and vaccine development. In the diagnosis of immune diseases, abnormalities in TCR diversity or the proliferation of specific TCR clones can serve as diagnostic markers.
Provide Insights for Tumor Immunotherapy Research
A study published in Immunother Cancer introduced an innovative method for directly identifying new antigen-specific TCRs from tumor samples using high-throughput single cell sequencing. Compared to traditional techniques such as T cell cloning and Sanger sequencing, this method significantly reduces the time and labor requirements, presenting a more efficient tool for tumor immunotherapy research and clinical applications.
Research background
The research focused on the isolation of new antigen-specific TCRs from tumor specimens of melanoma and colorectal cancer patients. Tumor-infiltrating T cells were isolated and processed for further analysis. CD14⁺ monocytes were purified from autologous peripheral blood mononuclear cells and differentiated into dendritic cells (DCs). Through a series of screening and single cell sequencing steps, new antigen-specific TCR sequences were identified. The full-length TCR sequences were then synthesized and cloned into retrovirus expression vectors for functional verification.
Experimental method
- Isolation of tumor-infiltrating T cells: Tumor-infiltrating T cells were isolated from tumor specimens of patients with melanoma and colorectal cancer, some of which were used for long-term TIL culture, and a few were made into single-cell suspension, frozen after treatment, and sorted by flow cytometry after resuscitation culture.
- Identification of new antigen-specific TCR sequence by single cell sequencing: CD14⁺ monocytes were purified from autologous peripheral blood mononuclear cells and cultured into dendritic cells (DCs). In order to reduce the number of peptide pool (PPs) and tandem mini-gene (TMG) in subsequent single cell analysis, melanoma samples were screened and determined, and colorectal cancer samples were directly screened because of the small number of cells.
- Verification of new antigen-specific TCR: The full-length TCR sequence was synthesized and cloned into retrovirus expression vector, and the virus supernatant was obtained by transfection of 293GP cells, which was used to transduce healthy donor PBMCs, co-cultured with DCs loaded with new antigen, and the secretion level of IFN-γ was determined by ELISA.
Experimental result
- Identification of new antigen-specific TCR from melanoma specimens: After tumor cells were co-cultured with DCs expressing TMG-1, single cell sequencing found that 26 single cells were highly expressing IFN-γ, and six different TCR were identified. After synthesis, autologous T cells were transduced and new antigens recognized by them were identified.
- Isolation of new antigen-specific TCR from colorectal cancer specimens: T cells that responded to PPs were screened out, and some shared and unique TCR were found after stimulation, and some could recognize mutant peptides. Single cell analysis was conducted directly. After stimulation, it was found that some single cells expressed IFN-γ or IL-2 at a high level, and various TCRs were identified, and some of them could recognize mutant peptides.
- The proportion of new antigen-specific TCR clones in tumor infiltrating T cells is small: In all tumor specimens, the frequency of new antigen-specific TCR clones in tumor infiltrating T cells is low, and the frequency of new antigen-reactive T cells detected by this method is lower than that by the previous low-flux single-cell method.
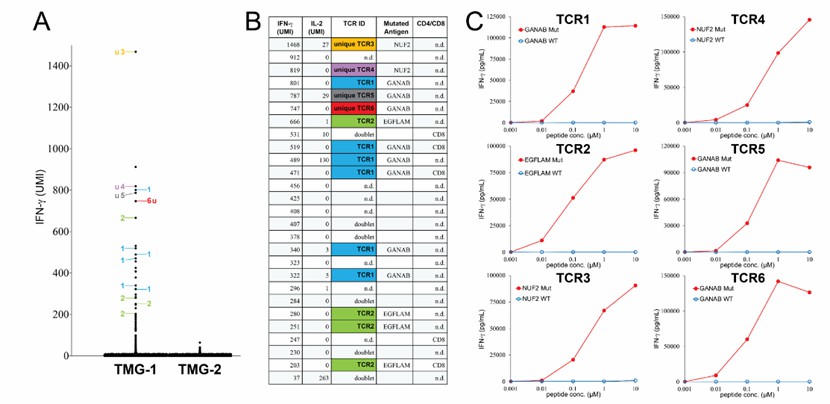 Neoantigen-specific TCRs from a metastatic tumor resected from one patient with melanoma (Lu et al., 2021)
Neoantigen-specific TCRs from a metastatic tumor resected from one patient with melanoma (Lu et al., 2021)
Discussion
- The new method can identify new antigen-specific TCR from melanoma and colorectal cancer samples, which saves time and effort compared with the previous long-term TIL culture method, but it has some limitations such as high cost and the need for a large number of tumors to infiltrate T cells.
- Compared with other methods for identifying TCR sequences, this method is relatively fast and efficient, but it still needs to be improved.
- In tumor infiltrating T cells, the ratio of new antigen to tumor reactive TCR is low, which may suggest that only a small number of T cells in tumor microenvironment can kill tumor cells, which needs large-scale research and verification.
- In the future, other markers (such as CD39 and CD103) will be tested to enrich new antigen-reactive T cells, and the improvement of single cell sequencing technology is expected to improve the efficiency of this method.
New Strategy for Personalized Treatment for Tumor
In one paper (publish in Frontiers in Immunology), Qualcomm quantity TCR sequencing technology was used for the first time to comprehensively analyze the characteristics of T cell receptor library in peripheral blood, tumor tissues and paired sentinel lymph nodes of patients with cervical cancer, revealing its potential value in immune monitoring and prognosis evaluation, providing an important basis for in-depth understanding of the immune status of patients with cervical cancer and providing a new direction for follow-up research.
Research background
- Cervical cancer is the fourth most common cancer among women in the world. Persistent infection with high-risk human papillomavirus (HPV) is the main cause. Host immunity, especially T-cell immunity, plays an important role in anti-tumor or anti-HPV infection mechanism, but there are few studies on the relationship between T-cell immune response and the pathogenesis of cervical cancer.
- T cell receptor library based on deep sequencing can be used as a biomarker of immune response in cancer patients, but the characteristics and prognostic significance of T cell receptor library in cervical cancer patients are still unclear.
Experimental method
- RNA extraction and TCR sequencing: Peripheral blood mononuclear cells (PBMCs) were separated by density gradient centrifugation, tissues and PBMC samples were lysed with TRIzol reagent, total RNA was extracted, TCRβ library was constructed by semi-nested PCR amplification and 5' rapid amplification of cDNA terminal (RACE), and sequenced on Hiseq 2500 platform.
- Sequencing data analysis: Filter low-quality sequences, compare the sequences with TCR reference genes by BLAT, determine CDR3 amino acid sequence, calculate sample diversity (Shannon's entropy index) and similarity (TCR library overlap rate), identify disease-related clones, and make statistical analysis.
Experimental result
- Basic information and sequencing data of patients: The samples were sequenced by Qualcomm TCR, and a large number of amino acid sequences of TCRβ CDR3 were obtained, and the use of V and J genes was determined. The clinical characteristics of patients with cervical cancer were diverse.
- Diversity of TCR pool in peripheral blood: The number of unique amino acid sequences and Shannon's entropy index of TCR pool in cervical cancer patients were significantly lower than those in CIN patients and healthy women, and the diversity of TCR pool in peripheral blood was lower than that in early patients, suggesting that the diversity of TCR pool in peripheral blood gradually decreased during the occurrence and development of cervical cancer.
- Usage of V and J genes in peripheral blood TCR bank: There were significant differences in the frequency of using some V genes among the three groups. Healthy women were different from cervical cancer and CIN patients, but there was no significant difference in the use of J gene and the distribution of CDR3 length.
- Similarity of TCR pool in peripheral blood: The overlapping rate of TCR pool in cervical cancer patients is higher than that in CIN patients and healthy women, and the overlapping rate between cervical cancer patients and CIN patients is higher than that in healthy women, indicating that the TCR pool in peripheral blood of cervical cancer patients is more similar to that in CIN patients.
- Disease-related clones: The disease-related clones unique to patients with cervical cancer and CIN were identified, and the CDR3 region has specific amino acid characteristics, which may constitute disease-specific markers.
- Diversity of TCR pool in tumor tissue and paired sentinel lymph nodes: The diversity of TCR pool in tumor tissue of cervical cancer patients is lower than that in paired sentinel lymph nodes, and the use of some V genes is different. Age has no effect on the diversity of TCR pool in both of them.
- The relationship between TCR bank and the prognosis of cervical cancer: the small number of clones in sentinel lymph nodes is related to the poor prognosis of cervical cancer patients, and there is no significant difference between the related indexes in tumor tissues and the prognosis.
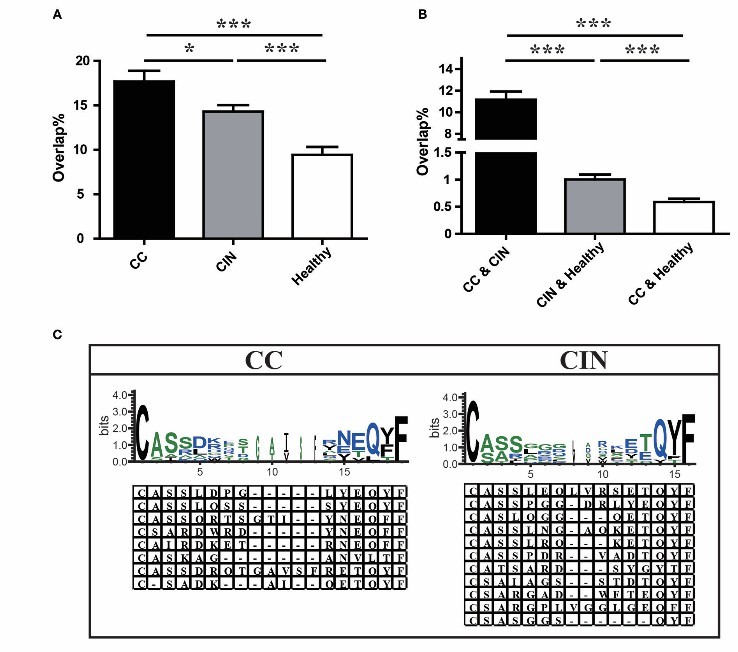 Comparison of clonotypes in peripheral blood TCR repertoire in different groups (Cui et al., 2018)
Comparison of clonotypes in peripheral blood TCR repertoire in different groups (Cui et al., 2018)
Discussion
- In this study, the characteristics of TCR pool in different samples of cervical cancer patients were comprehensively described for the first time. It was found that the diversity of TCR pool in peripheral blood decreased gradually, which may be related to T cell depletion, and it can be used as a potential biomarker to monitor the immune changes of cervical cancer patients.
- The high similarity of TCR library between cervical cancer patients and CIN patients may be attributed to the similar immune environment caused by long-term HPV infection.
Reveal Potential Value in Immune Monitoring and Prognosis Evaluation
The low diversity of TCR library in tumor tissue may reflect the specific response of oligoclonal T cells to tumor-associated antigens. The number of clones in sentinel lymph nodes is related to prognosis, and its overlapping clones may be effector memory T cells, which plays an important role in monitoring tumor recurrence, but further research is needed to determine its exact impact.
Reearch background
- The recognition of tumor-specific mutation (new antigen) by T cells plays a key role in cancer immunotherapy, but the identification of new antigen-reactive T cells is difficult because of the patient specificity and complexity of the antigen.
- Traditional methods have limitations, such as relying on algorithm prediction, difficulty in verification, and lack of tumor cell lines as experimental reagents.
- Phenotypic marker-based methods can be used to identify tumor reactive T cells, but more effective strategies are needed.
Experimental method
- TCR gene was cloned from TIL sorted by FACS, transduced into 293gp cells, co-cultured with autologous dendritic cells with pulsed mutant or wild-type peptides, and T cell activation was detected by IFN-γ ELISA.
- Single cell analysis (CITE-seq and TCR-seq): Some fresh tumor samples were used for TIL culture, and the rest were made into single cell suspensions, which were analyzed by CITE-seq, including cell surface protein staining, sorting, library preparation and sequencing. The data were analyzed by special software to determine the characteristics of new antigen-reactive T cells.
- CDR3β deep sequencing: PBL samples obtained during tumor resection were sent for testing, and the sequence frequency of TCRβ CDR3 was analyzed.
- TCR selection and evaluation: T cells were selected according to the expression of CD39 protein, and high-frequency clones were selected for different groups of clones. The candidate TCR was synthesized and transduced into autologous PBLs, and co-cultured with mutant peptides. The reactivity was detected by IFN-γ ELISA.
Experimental result
- Isolation of new antigen-reactive T cells from NSCLC Tiles: NSCLC Tiles were screened by traditional methods, and 11 kinds of mutation-reactive TCR were identified to determine their new antigen specificity; CITE-seq analysis of T cells in fresh tumor digests showed that new antigen-reactive T cells had certain distribution characteristics in cell surface protein expression and transcriptome analysis.
- High expression of CD39 and CXCL13 in new antigen-reactive CD8 T cells: Comparing the protein and gene expression of new antigen-reactive CD8 T cells with other CD8 T cells, we found a number of differentially expressed genes and protein, in which CD39 and CXCL13 were significantly enriched in new antigen-reactive cells, and the new antigen-reactive T cells had a depleted phenotype.
- New antigen-reactive CD8⁺ T cells can be identified based on the characteristics of high-frequency TCR clone, co-expression of CD39 protein and CXCL13 mRNA: In patient samples, the proportion of CD39 cells in different patients is different, and the frequency of CXCL13 expression cells is also different; The specific clonal TCR was selected for evaluation, and it was found that some TCR had new antigen reactivity, which indicated that this feature could be used to identify new antigen and new antigen reactivity TCR. Most new antigen-reactive CD8⁺ T cells express CD39 and CD103, and the expression of PD-1 is inconsistent, and the expression of TIM3 is also inconsistent.
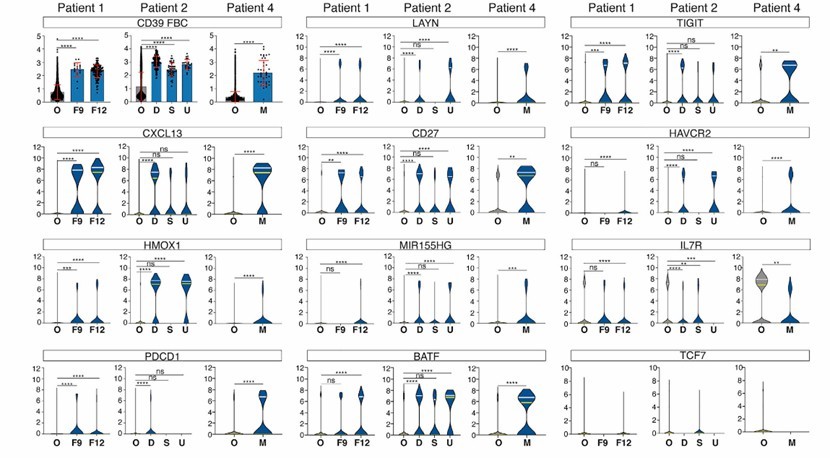 Analysis of gene and protein expression on neoantigen-reactive T cells at the clonotype level (Hanada et al., 2022)
Analysis of gene and protein expression on neoantigen-reactive T cells at the clonotype level (Hanada et al., 2022)
Discussion
- The phenotypic characteristics developed in this study can quickly identify new antigen-reactive T cells by directly using TILs in fresh tumor specimens, which overcomes some limitations of traditional methods.
- The expression of CD39 and CXCL13 can recognize CD8⁺ and CD4⁺ new antigen-reactive T cells, but the frequency may be underestimated, because other types of tumor-associated antigens or new antigens may be missed by screening methods.
- CXCL13 is highly expressed in many cancers, and its high frequency expression in new antigen-reactive TILs is related to other research results, but its low frequency in TILs requires better screening or enrichment methods.
- Although the TCR identified in this study has new antigen reactivity, the epigenetic status of cells may affect their therapeutic potential. In the future, cloned TCR can be used to genetically modify T cells for adoptive cell therapy.
- The results of this study need to be verified in more tumor types, and the influence of current immunotherapy on this feature needs to be determined. This feature has potential value in identifying new antigen-reactive clones and is helpful to develop effective cancer immunotherapy.
The advantages of TCR sequencing are high sensitivity and Qualcomm, which can detect low-frequency TCR clones and quickly analyze a large number of samples. However, facing the challenges of complex data interpretation and high sample preparation requirements, professional bioinformatics knowledge and standardized processes are needed.
References
- AF Sanroman, K Joshi, L Au, B Chain and S Turajlic. "TCR sequencing: applications in immuno-oncology research." Immuno-Oncology and Technology (2023) https://doi.org/10.1016/j.iotech.2023.100373
- Yongchen Lu, Zhili Zheng, Frank J Lowery, Jared J Gartner, Todd D Prickett, and Steven A Rosenberg. "Direct identiffcation of neoantigen-speciffc TCRs from tumor specimens by high- throughput single- cell sequencing." Journal for Immuno Therapy of Cancer (2021):e002595. doi:10.1136/jitc-2021-002595
- Jinhua Cui, Kairong Lin, Songhua Yuan and Wei Luo. "TCR Repertoire as a Novel Indicator for Immune Monitoring and Prognosis Assessment of Patients With Cervical Cancer." Frontiers in Immunology (2018) doi: 10.3389/fimmu.2018.02729
- Ken-ichi Hanada, Chihao Zhao, Raul Gil-Hoyos and James C Yang. "A phenotypic signature that identifies neoantigenreactive T cells in fresh human lung cancers." Cancer Cell (2022) 479–493. https://doi.org/10.1016/j.ccell.2022.03.012
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

 Schematic principle of TCR sequencing (Sanroman et al., 2023)
Schematic principle of TCR sequencing (Sanroman et al., 2023) Neoantigen-specific TCRs from a metastatic tumor resected from one patient with melanoma (Lu et al., 2021)
Neoantigen-specific TCRs from a metastatic tumor resected from one patient with melanoma (Lu et al., 2021) Comparison of clonotypes in peripheral blood TCR repertoire in different groups (Cui et al., 2018)
Comparison of clonotypes in peripheral blood TCR repertoire in different groups (Cui et al., 2018) Analysis of gene and protein expression on neoantigen-reactive T cells at the clonotype level (Hanada et al., 2022)
Analysis of gene and protein expression on neoantigen-reactive T cells at the clonotype level (Hanada et al., 2022)