What is Whole Plasmid Sequencing?
Whole Plasmid Sequencing (WPS) also referred to as full plasmid sequencing or complete plasmid sequencing, involves the comprehensive sequencing of an entire plasmid DNA molecule. Plasmids, which are circular DNA molecules commonly found in bacteria, play significant roles in genetic engineering, molecular cloning, and vaccine development. By providing the complete genetic sequence of plasmids, WPS is crucial for ensuring plasmid functionality and identifying potential gene mutations or variations in resistance genes.
Unlike traditional fragmented sequencing, WPS enables the direct determination of the entire DNA sequence of a plasmid without necessitating fragmentation into multiple segments prior to sequencing. This approach improves efficiency and reduces sequencing errors, like misassemblies and information loss.
Currently, WPS predominantly utilizes two mainstream high-throughput sequencing technologies: Next-Generation Sequencing (NGS) and Nanopore Sequencing. NGS is renowned for its high accuracy but may encounter challenges when dealing with large or complex plasmids. Conversely, Nanopore Sequencing provides long-read data, facilitating rapid and comprehensive plasmid analysis, especially advantageous for larger plasmids or those containing repetitive sequences.
Why is Whole Plasmid Sequencing Important?
Whole plasmid sequencing is an indispensable tool in the life sciences, profoundly influencing areas such as antibiotic resistance, genetic engineering, and clinical diagnostics. Its importance is underscored by multiple key advantages which include:
1. Comprehensive Plasmid Analysis:
Plasmids often carry important genes, such as those that provide antibiotic resistance or produce toxins. Conventional sequencing techniques often fall short in delivering complete plasmid sequences. WPS addresses this gap by providing exhaustive plasmid genome data, essential for a comprehensive understanding of plasmid-mediated functions.
2. Unraveling Antibiotic Resistance Mechanisms:
Plasmids are central to the horizontal transfer of antibiotic resistance genes among bacteria. Utilizing WPS, researchers can meticulously track the spread of these resistance genes, illuminating the underlying mechanisms. Such insights are crucial for monitoring antibiotic resistance trends and formulating effective therapeutic strategies.
3. Efficient Assembly of Complex Plasmids without a Reference:
Contrasting with traditional sequencing methodologies, WPS does not necessitate a reference genome for assembly. This is especially helpful for assembling complex plasmids with high GC content, repetitive sequences, or large sizes, leading to better assembly quality.
4. Cost and Time Efficiency:
When compared to conventional sequencing, WPS offers significant reductions in both time and cost. Through automated workflows, plasmid sequencing projects can be completed in mere days, highlighting a distinct cost advantage, particularly for larger plasmids.
5. Broad Applications Across Diverse Fields:
The utility of WPS extends beyond fundamental research, encompassing extensive applications in clinical diagnostics, vaccine development, and genetic engineering. By enabling the rapid identification of resistant strains and the optimization of genetic vectors, WPS propels advancements in precision medicine and synthetic biology.
In conclusion, whole plasmid sequencing furnishes an efficient and precise technological bedrock for plasmid research, wielding significant implications for antibiotic resistance surveillance, clinical applications, and innovations in genetic engineering.
CD Genomics offers a comprehensive range of plasmid sequencing services, utilizing advanced sequencing technologies for accurate and customized analysis of plasmid DNA.
How Many Gene Sequences Are in a Plasmid?
Plasmids are small DNA molecules that replicate independently in bacterial cells. They play a crucial role in providing various traits, such as antibiotic resistance, but are not involved in essential cell growth and reproduction. These circular DNA structures can encode a variety of proteins and RNAs, endowing plasmids with specific functionalities such as antibiotic resistance, heavy metal tolerance, and toxin production.
Genetic Composition and Functional Categories
The diversity in gene sequences within a plasmid is influenced by its size and the functional capacities it supports. Smaller plasmids may encompass only a few genes, whereas larger ones can include multiple genes along with associated regulatory elements. Plasmid-encoded genes generally categorize into:
- Resistance Genes: These primarily confer antibiotic resistance, allowing bacteria to persist in environments with antibiotic presence. Such genes are among the most frequently encountered on plasmids.
- Replication and Regulatory Genes: These are vital for initiating replication and maintaining and regulating the plasmid. They encompass replication origins, promoters, and terminators to ensure stable plasmid replication and transmission.
- Foreign Gene Insertion Sites: In genetic engineering, plasmids often incorporate multiple cloning sites (MCS) to facilitate the insertion of foreign genes and other genetic elements.
- Toxin Genes: Some plasmids carry genes encoding toxins or antitoxins, impacting the pathogenic potential of the host bacterium.
Plasmid Size and Gene Content
Plasmid sizes correlate with the number of genes they harbor. Below is a summary of common plasmids, their sizes, and typical gene content:
| Plasmid Name |
Size (kb) |
Number of Genes |
Common Function |
| pUC19 |
2.7 |
2-3 |
Cloning vector, carries ampicillin resistance gene |
| pBR322 |
4.4 |
3-4 |
Cloning vector, includes ampicillin and tetracycline resistance genes |
| pET-28a |
5.8 |
4-5 |
Expression vector for protein systems |
| pGEM-T |
3.0 |
3-4 |
PCR product cloning |
| pBluescript SK+ |
3.0 |
3-4 |
DNA sequencing and subcloning |
| pACYC177 |
4.0 |
2-3 |
Contains chloramphenicol resistance gene |
| pLysS |
3.0 |
3-4 |
Expression vector controlling expression through T7 lysozyme |
| pET-32a |
5.8 |
4-5 |
Includes fusion tag for protein purification |
| pSTBlue-1 |
3.0 |
3-4 |
Sequencing and mutagenesis |
Note: Gene numbers might vary slightly due to plasmid variants or modifications.
Gene Content by Plasmid Size:
- Small Plasmids (< 5 kb): Typically possess one or a few functional genes, such as those seen in the pUC series (~2.5 kb), which include antibiotic resistance genes and MCS for efficient genetic engineering experiments.
- Medium-sized Plasmids (5 - 30 kb): These plasmids, like pBR322 (~4.3 kb), include functions for molecular cloning and antibiotic selection, while the pET series (3-6 kb) provides promoters for recombinant protein expression.
- Large Plasmids (> 30 kb): Such plasmids may contain a wide range of genes and are often studied for their role in resistance research. R plasmids provide significant survival advantages to host bacteria under antibiotic pressure.
- F Plasmids (> 100 kb): Represent typical bacterial conjugative plasmids, with genomes reaching up to 120 kb, carrying genes vital for bacterial mating, replication, and inter-bacterial transfer, involving multiple regulatory sequences.
Through the detailed analysis of gene content and function in plasmids, researchers gain comprehensive insights into their biological roles, fostering advancements in antibiotic resistance studies, molecular genetics, and biotechnological applications.
Direct Colony to Whole Plasmid Sequencing
In contemporary molecular biology research, whole plasmid sequencing has significantly enhanced the efficiency and precision with which complete plasmid genomes are obtained directly from bacterial colonies. Traditionally, the extraction and sequencing of plasmids required the isolation of monoclonal colonies for DNA extraction, followed by analysis using standard NGS or Sanger sequencing techniques. This conventional approach is not only labor-intensive and time-consuming but also poses limitations in success rate and accuracy, particularly for plasmids lacking reference sequences or with intricate structures—such as large plasmids, those with high GC content, or those containing repetitive regions.
Whole plasmid sequencing, employing technologies such as nanopore sequencing with long-read capabilities or other cutting-edge whole genome sequencing methodologies, markedly improves both the efficiency and accuracy of plasmid sequencing. This technology facilitates the direct acquisition of complete plasmid genomes from bacterial cultures and addresses the challenges faced by traditional techniques in the assembly of complex plasmids without the need for reference sequences.
How is Whole Plasmid Sequencing Done?
Whole plasmid sequencing employs cutting-edge genomic technologies to obtain a complete and precise sequence of plasmid DNA. Unlike traditional sequencing methods, WPS facilitates the assembly of complex plasmid structures without the need for a reference sequence. Herein, we delve into the procedural steps of WPS and the technologies that power this innovative approach.
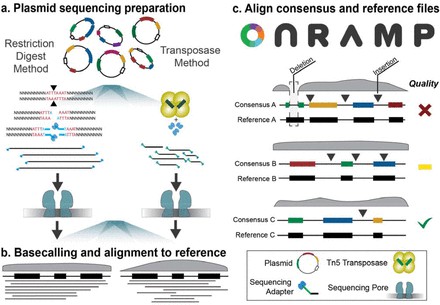 Plasmid library preparation and consensus sequence generation using On-Ramp. (Mumm, Camille, et al., bioRxiv, 2022)
Plasmid library preparation and consensus sequence generation using On-Ramp. (Mumm, Camille, et al., bioRxiv, 2022)
Sample Preparation and Quality Assessment
The initial phase of WPS involves extracting plasmid DNA from bacterial colonies, ensuring that samples meet stringent quality standards. Standard extraction procedures, such as alkaline lysis or commercially available plasmid extraction kits, are utilized. Once extracted, the plasmid DNA undergoes quality checks, including assessments of concentration, purity, and integrity, typically via spectrophotometry or agarose gel electrophoresis. Ensuring high-quality DNA is crucial for accurate sequencing outcomes.
Sample Processing and DNA Fragmentation
Following extraction and quality assessment, DNA samples are prepared for sequencing by fragmentation. Traditional short-read sequencing techniques, like those platformed by Illumina, utilize enzymatic or mechanical means to cleave DNA into manageable fragments. In WPS, Tn5 transposase is often used for DNA fragmentation, efficiently cleaving long DNA chains while appending adaptor sequences to the fragments’ ends. These adaptors include essential “barcode” sequences for polymerase chain reaction (PCR) amplification and sequencing.
The integration of barcodes is pivotal, as it facilitates sample differentiation within high-throughput sequencing platforms, allowing for seamless separation and analysis of mixed sample origins following sequencing.
Library Construction and Adaptor Ligation
Post-transposase treatment, DNA fragments are ligated with sequencing adaptors, forming the sequencing library. Adaptors are sequences necessary for library recognition and amplification in sequencing platforms. PCR amplification is applied to ensure a sufficient quantity of DNA fragments is present, providing the necessary depth for comprehensive and accurate sequencing.
Sequencing Execution
Upon library construction, sequencing is conducted using high-throughput platforms such as Illumina, PacBio, or Oxford Nanopore:
- Illumina Sequencing: Predominantly utilized for its accuracy, albeit providing shorter read lengths. Techniques like bridge PCR and single-molecule detection facilitate the efficient acquisition of plasmid sequences.
- PacBio and Oxford Nanopore Sequencing: These platforms offer long-read capabilities advantageous for resolving complex plasmid structures. Long reads enhance assembly performance, particularly in repetitive or GC-rich regions. Notably, Oxford Nanopore can directly sequence long strands of DNA, thus enhancing sequencing speed and genome coverage.
Data Processing and Genome Assembly
The sequencing outputs undergo rigorous data processing, including quality control, filtering, and error correction:
- Data Filtering: Low-quality sequences and potential contaminants are removed to maintain the integrity of the dataset.
- Error Correction: Software tools such as Medaka are employed to rectify sequencing inaccuracies, critical for high-fidelity genome assembly.
- Assembly: Fragmented sequencing reads are assembled into a complete plasmid genome using assemblers like Flye, SPAdes, or miniasm, each leveraging long-read attributes for robust assembly accuracy.
Result Interpretation and Annotation
The assembled plasmid genome undergoes comparative analysis with established databases to identify functional genes, regulatory regions, and potential mutation sites. This comprehensive annotation allows researchers to discern plasmid functionalities, such as antibiotic resistance determinants, toxin production genes, or metabolic pathway components.
Advanced platforms also facilitate variant analysis, pinpointing mutations, deletions, insertions, and rearrangements—offering insights into plasmid evolution across diverse environments, pertinent to pharmaceutical innovation and clinical diagnostics.
Automation and Output Integration
A hallmark of WPS is its streamlined automation, significantly reducing manual intervention and enhancing throughput. Automated pipelines efficiently translate raw sequencing data into actionable insights. Visual outputs allow researchers to readily interpret results using alignment and annotation software, paving the way for further biological experimentation, including gene functional studies and antimicrobial resistance assays.
In summary, Whole Plasmid Sequencing revolutionizes plasmid analysis through its high efficiency, precision, and applicability across multiple domains such as resistance monitoring, clinical applications, and genetic engineering, affirming its position as an indispensable tool in contemporary life sciences.
How Long Does Whole Plasmid Sequencing Take?
The duration required for whole plasmid sequencing is contingent upon the employed technology and the intricacy of the plasmid itself:
- Next-Generation Sequencing: Traditional NGS methodologies typically require several hours to a full day to complete the sequencing process. Subsequent data analysis may extend over several days. This technology is well-suited for smaller plasmids with relatively simple sequences.
- Nanopore Sequencing: In comparison to NGS, nanopore sequencing offers a considerably faster sequencing rate, particularly advantageous for large or complex plasmids. Data generation through nanopore sequencing can be accomplished within a matter of hours. When utilizing portable devices such as the MinION, small-scale sequencing projects can generally be completed, including preliminary analyses, within a single day.
In summary, nanopore sequencing presents a significant time advantage in whole plasmid sequencing, making it especially suitable for applications where rapid acquisition of plasmid sequences is imperative.
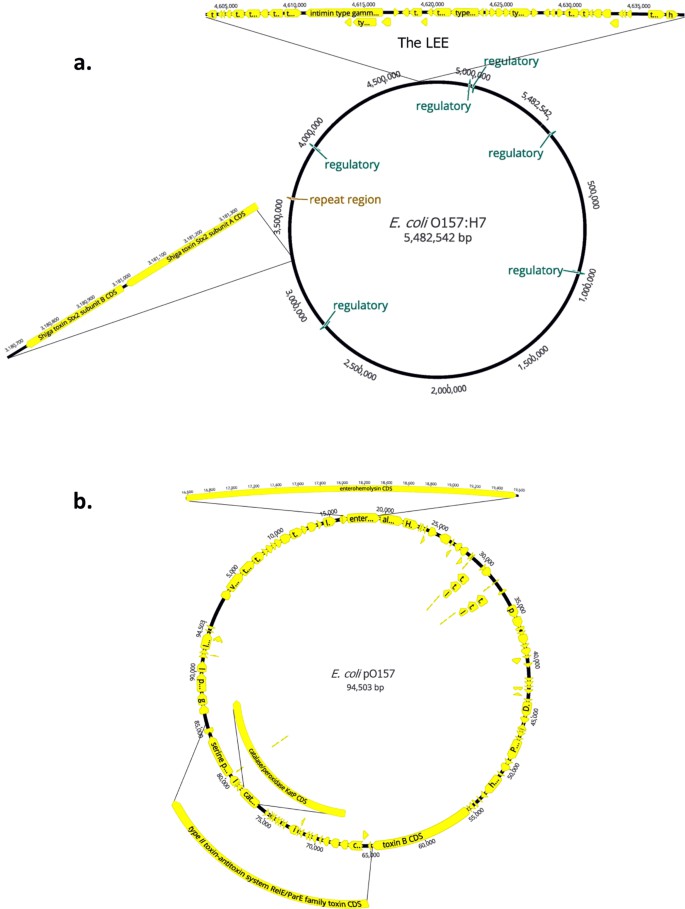 Annotation of the MinION assembly of Escherichia coli. (Taylor, T.L. et al. Sci Rep, 2019)
Annotation of the MinION assembly of Escherichia coli. (Taylor, T.L. et al. Sci Rep, 2019)
Difference Between Whole Plasmid Sequencing and Sanger Sequencing
Whole plasmid sequencing offers a range of advantages over traditional Sanger sequencing, particularly with respect to sequencing accuracy, cost-effectiveness, efficiency, and scope of application. Here, we delineate the primary distinctions between whole plasmid sequencing and Sanger sequencing, providing a tabular comparison across several dimensions to enhance clarity and comprehension.
| Comparison Item |
Sanger Sequencing |
Whole Plasmid Sequencing |
| Read Length |
Short reads (typically 500-1000 base pairs per read) |
Long reads (Oxford Nanopore can achieve several thousand to tens of thousands of base pairs) |
| Applicable Plasmid Size |
Suitable for small plasmids (generally <10 kb); challenging for larger plasmids |
Suitable for plasmids of all sizes, including large plasmids (>100 kb) |
| Dependency on Reference Sequence |
Highly reliant on reference sequences, requiring primer design for each plasmid |
Independent of reference sequences, capable of de novo assembly without primer design |
| Sequencing Speed |
Slow, requiring multiple sequencing cycles (from days to weeks) |
Fast, typically producing results within 2-5 working days |
| Error Rate |
Higher error rate, particularly in complex regions (e.g., high GC content and repetitive regions) |
Lower error rate, with long reads reducing low-quality sequencing data |
| Data Processing |
Requires manual examination of chromatograms, significant human intervention |
Highly automated, with assembly and analysis processes reducing human error |
| Cost |
High cost, especially for large plasmids, with typically high per kb sequencing fees |
Lower cost, particularly for large plasmids, with costs diminishing as plasmid size increases |
| Scope of Application |
Primarily for short, known reference sequence plasmid sequencing |
Suitable for varied plasmids, particularly complex ones lacking reference sequences, or scenarios requiring high-throughput sample processing |
| Handling of Repetitive Sequences |
Difficulties handling repetitive and high GC content regions, potentially leading to assembly failure |
Superior performance in assembling complete genomes through repetitive and high GC regions |
| Sequencing Platforms |
Based on conventional fluorescent labeling technologies, employing ABI or Applied Biosystems platforms |
Utilizes modern high-throughput sequencing technologies, with platforms such as Oxford Nanopore and PacBio prevalent |
| Assembly and Stitching |
Each fragment sequenced and manually stitched, may require iterative cycles |
Fully automated stitching, with long reads facilitating the handling of long fragments and complex structures |
| Fields of Application |
Predominantly used for precise gene sequencing, mutation detection, PCR product sequencing, etc. |
Extensively applied in plasmid, genome assembly, antibiotic resistance gene monitoring, synthetic biology, etc. |
Key Comparative Insights:
- Read Length: Sanger sequencing is limited to shorter read lengths (typically 500-1000 base pairs), while whole plasmid sequencing is capable of producing long-read data, enabling the coverage of more extensive genomic regions, and is particularly suited for comprehensive sequencing of large plasmids.
- Dependency on Reference Sequence: Sanger sequencing necessitates the design of specific primers for target plasmids and relies on known reference sequences for sequencing. In contrast, whole plasmid sequencing circumvents the need for reference sequences by facilitating de novo assembly, making it especially advantageous for dealing with unknown or novel plasmids.
- Ability to Handle Complex Structures: Sanger sequencing encounters substantial limitations in regions of repetition or high GC content or in plasmids with unique secondary structures, often resulting in assembly challenges. Whole plasmid sequencing, especially supported by long-read technologies like Oxford Nanopore, effectively addresses these challenges, yielding precise whole plasmid sequences.
- Cost and Speed: Sanger sequencing is generally expensive, particularly for large plasmids, and involves protracted sequencing durations. Whole plasmid sequencing offers a cost-effective solution for large plasmids, rapidly delivering comprehensive genomic information.
- Automation and Data Processing: Sanger sequencing often requires manual intervention, particularly in chromatogram interpretation and data assembly. Conversely, whole plasmid sequencing automates data processing, enabling efficient assembly and analysis while minimizing human error.
In summary, whole plasmid sequencing significantly surpasses Sanger sequencing not only in terms of cost, speed, and accuracy but also in its capacity to handle complex plasmids, plasmids lacking reference sequences, and the demands of large-scale, high-throughput sequencing. This provides a more flexible and efficient solution for a wide array of biomedical and synthetic biology applications.
If you would like to learn more about plasmids and whole plasmid sequencing, you can read the following articles:
Unraveling Plasmids: A Comprehensive Guide
Plasmid Detection and Complete Plasmid DNA Sequencing
Plasmid Fact Sheet: Definition, Structure and Application
Exploring Plasmid Extraction: Techniques and Key Considerations
References:
- Mumm, Camille, et al. "On-Ramp: A tool for rapid, multiplexed validation of plasmids using nanopore sequencing." bioRxiv (2022): 2022-03. doi: https://doi.org/10.1101/2022.03.15.484480
- Bai, Xingjian, et al. "Prevalence of errors in lab-made plasmids across the globe." bioRxiv (2024): 2024-06. doi: https://doi.org/10.1101/2024.06.17.596931
- Wick, Ryan R., et al. "Recovery of small plasmid sequences via Oxford Nanopore sequencing." Microbial genomics 7.8 (2021): 000631. https://doi.org/10.1099/mgen.0.000631
- Zhao, Wenxuan, et al. "Oxford nanopore long-read sequencing enables the generation of complete bacterial and plasmid genomes without short-read sequencing." Frontiers in Microbiology 14 (2023): 1179966. https://doi.org/10.3389/fmicb.2023.1179966
- Kopotsa, Katlego, John Osei Sekyere, and Nontombi Marylucy Mbelle. "Plasmid evolution in carbapenemase‐producing Enterobacteriaceae: a review." Annals of the New York Academy of Sciences 1457.1 (2019): 61-91. https://doi.org/10.1111/nyas.14223
- Garcillán-Barcia, M. Pilar, Santiago Redondo-Salvo, and Fernando de la Cruz. "Plasmid classifications." Plasmid 126 (2023): 102684. https://doi.org/10.1016/j.plasmid.2023.102684
- Carattoli, Alessandra, and Henrik Hasman. "PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS)." Horizontal gene transfer: methods and protocols (2020): 285-294. https://doi.org/10.1007/978-1-4939-9877-7_20


 Sample Submission Guidelines
Sample Submission Guidelines
 Plasmid library preparation and consensus sequence generation using On-Ramp. (Mumm, Camille, et al., bioRxiv, 2022)
Plasmid library preparation and consensus sequence generation using On-Ramp. (Mumm, Camille, et al., bioRxiv, 2022) Annotation of the MinION assembly of Escherichia coli. (Taylor, T.L. et al. Sci Rep, 2019)
Annotation of the MinION assembly of Escherichia coli. (Taylor, T.L. et al. Sci Rep, 2019)