In the fields of molecular biology and genetic engineering, plasmids serve as vital genetic vectors widely utilized in gene cloning, expression, modification, and therapeutic research. This article aims to explore the multifaceted aspects of plasmids, including their definition, historical discovery, structural characteristics, functions, and distinctions from DNA vectors.
What is a Plasmid?
Definition of Plasmids:
Plasmids are small, circular DNA molecules. They exist separately from the main DNA in cells and can copy themselves. They are predominantly found in bacteria and archaea, but can also be identified in certain eukaryotic organisms, such as yeast. Due to their simple structure, clear functionality, and ease of manipulation, plasmids have become widely utilized in the realms of genetic engineering, molecular biology, and biotechnology.
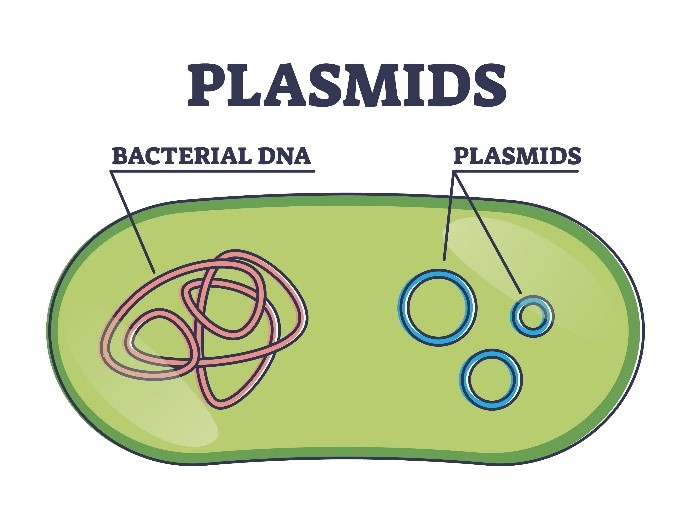
Fundamental Properties of Plasmids:
1. Copy Number
The copy number refers to the average quantity of plasmids per cell, which is determined by the plasmid's origin of replication. Based on their copy numbers, plasmids can be categorized into:
- High-Copy Number Plasmids: These can yield hundreds to thousands of copies per cell, such as the modified pUC plasmids (copy number 500-700). These plasmids are ideal for high-yield DNA preparation and gene expression experiments.
- Low-Copy Number Plasmids: Exhibiting only a few copies per cell (e.g., pMB1 plasmid, copy number 15-20), low-copy number plasmids are commonly employed for cloning large DNA fragments or toxic genes, minimizing resource consumption from the host and enhancing experimental success rates.
2. Replication Control Types
Plasmids can also be classified based on their replication control mechanisms:
- Relaxed Replication: These plasmids rely entirely on host-provided proteins for replication and can maintain high copy numbers, even under nutrient-limiting conditions, exemplified by pMB1.
- Stringent Replication: Such plasmids require the synthesis of their replication-associated proteins (e.g., RepA). Their replication capabilities are restricted when host protein synthesis is hindered, as is the case with pSC101 plasmids.
3. Incompatibility
Plasmid incompatibility occurs when two plasmids share the same origin of replication or similar regulatory systems, leading to competition for host resources during replication. Consequently, one plasmid can be excluded after successive amplifications. To mitigate concerns regarding plasmid competition in experiments, it is critical that different plasmids possess compatible origins of replication.
4. Selective Pressure
To ensure plasmid stability within host cells, antibiotics are often incorporated into growth media to apply selective pressure on plasmids carrying resistance genes. For example, by adding ampicillin (Amp) or kanamycin (Kan), only bacteria harboring the corresponding resistance genes are permitted to grow. This strategy not only facilitates the selection of successfully transformed cells but also increases plasmid concentrations.
Characteristics of Plasmids:
- Autonomous Replication: Plasmids contain an origin of replication (Ori) that enables them to harness the host's replication mechanisms for self-replication, thereby amplifying their copy numbers.
- Carriage of Additional Traits: Many plasmids harbor genes such as antibiotic resistance genes and toxin genes, which enhance the survival or adaptability of host cells.
- Incompatibility: Plasmids with identical origins of replication typically cannot coexist within the same cell due to their competition for the replication machinery, resulting in the exclusion of one plasmid.
- Transferability: Certain plasmids (e.g., F and R plasmids) possess the ability to transfer themselves between cells through conjugation, thus facilitating horizontal gene transfer.
How Was the Plasmid Discovered?
In the 1940s, scientists identified a cytoplasmic genetic factor capable of transferring between cells, leading to various hypotheses regarding its nature, including associations with parasites, symbionts, and genes. However, a definitive nomenclature for this entity remained elusive. It was not until 1952 that Joshua Lederberg began to clarify the classification of these cytoplasmic genetic factors and introduced the term "plasmid." This term is a composite of "cytoplasm" and the Latin suffix "-id," meaning "it," thereby designating any hereditary determinant that exists outside of a chromosome.
Despite this seminal contribution, Lederberg's definition did not gain immediate traction within the scientific community. A few years later, Élie Jacob and François Wollman proposed the term "episome," describing it as "a non-essential genetic element that can exist autonomously or integrate into chromosomes," which gained broader acceptance. Notably, Ester Lederberg's discovery in 1952 of the Fertility or F factor, which could integrate into the Escherichia coli chromosome under certain conditions, provided empirical support for the utility of this terminology.
A pivotal shift occurred in the 1960s when researchers discovered Resistance or R factors that could transfer between bacteria, yet unlike the F factor, R factors were unable to incorporate into the chromosome. This distinction facilitated the resurgence of the term "plasmid," which again became prominent within the scientific discourse.
By the 1970s, advancements such as restriction endonucleases, DNA ligases, and gel electrophoresis made the manipulation of DNA fragments feasible. The combination of these DNA fragments with plasmids or vector DNA frameworks led to the creation of recombinant DNA molecules, thus opening new avenues for the investigation of gene functions.
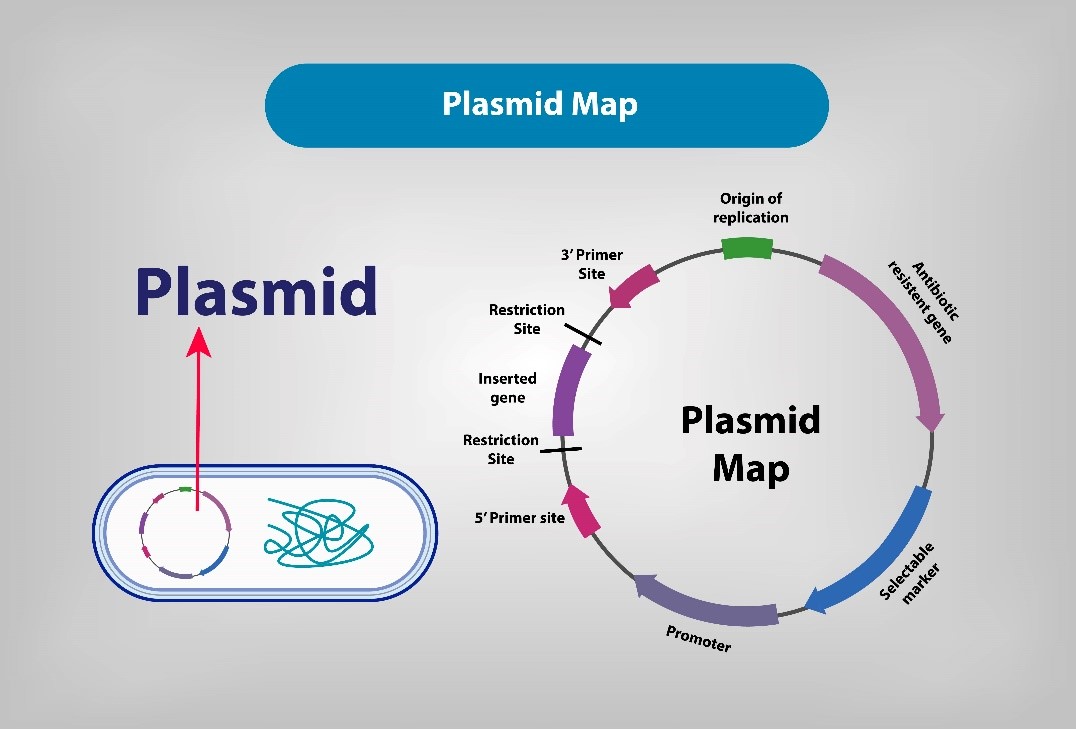
The Structure of Plasmids: How to Interpret a Plasmid?
Plasmids have different parts that help them work inside cells. Understanding these parts is important. Plasmids are not merely simplistic DNA carriers; rather, each structural component plays a pivotal role in processes such as replication, gene expression, cellular selection, and the insertion of foreign genes. By grasping these elements, researchers can more effectively employ plasmids in genetic cloning, gene expression studies, and various molecular biology experiments.
Plasmid sequencing methods, including Sanger sequencing and next-generation sequencing (NGS), are crucial for determining the precise nucleotide sequence of plasmids. These techniques help verify gene presence and orientation, detect mutations, and confirm successful cloning.
This section will integrate the main structural features of plasmids with methodologies for interpreting plasmid maps, thereby facilitating a deeper understanding of plasmid functionality.
Key Structural Features of Plasmids
1. Origin of Replication (ORI)
The ORI serves as the central feature of the plasmid, responsible for initiating autonomous replication. By activating the host cell's replication machinery, the ORI determines the plasmid's copy number and stability within the host. When selecting an appropriate ORI for experimental purposes, one must consider the plasmid's host range and compatibility with other plasmids.
2. Selectable Marker
Selectable markers are crucial for identifying successfully transformed host cells. Common markers include antibiotic resistance genes, such as the ampicillin resistance gene (Ampr), as well as fluorescent protein genes. In antibiotic-containing media, only cells that carry the plasmid will survive, thereby enabling efficient selection processes.
3. Multiple Cloning Site (MCS)
The MCS has special sites for adding new DNA. It is placed near the promoter, so the new gene can be expressed correctly.
4. Insert
The insertion of foreign DNA fragments into the plasmid via molecular cloning techniques is fundamental to the functionality of plasmids. The inserted fragments may include target genes, regulatory elements, or reporter genes.
5. Promoter Region
The promoter region governs the transcription of the target gene. Common promoters include sequences tailored for prokaryotic or eukaryotic hosts, such as the T7 promoter used in prokaryotic expression, and the CMV promoter frequently used in mammalian cell systems.
6. Primer Binding Site
Primer binding sites facilitate PCR amplification or sequencing validation, ensuring the accuracy of the plasmid sequence and the inserted fragments. These sites are typically located adjacent to the MCS, thus facilitating the design of universal primers.
7. Vector Backbone
The backbone of the plasmid constitutes its fundamental structure, incorporating the ORI, selectable marker gene, and MCS. The design aims for simplicity and stability, accommodating diverse experimental requirements.
 Analysis results of integrated ATAC-seq and RNA-seq results.
Analysis results of integrated ATAC-seq and RNA-seq results.
Interpreting a Plasmid Map
To understand how a plasmid works, scientists use a plasmid map. This map shows key parts of the plasmid, like where it starts copying itself and any genes it carries. Below are key steps for effectively interpreting a plasmid map:
1. Identify the Origin of Replication (ORI)
First, locate the ORI to understand the plasmid's copy number and host range. For experiments utilizing multiple plasmids, ensuring ORI compatibility is crucial to prevent instability caused by competitive replication mechanisms.
2. Examine Selectable Marker Genes
Confirm the presence of resistance genes on the plasmid, such as Ampr or Kanr, and select the corresponding antibiotic-containing media to ensure successful transformation selection.
3. Locate the Multiple Cloning Site (MCS)
Identify the restriction sites within the plasmid to determine optimal insertion points for foreign genes, ensuring that these insertions do not disrupt other functional areas.
4. Check the Expression System
Verify the inclusion of a promoter, ribosome binding site (RBS), and transcription termination signals. For gene expression experiments, it is critical to select a promoter type suitable for the intended host, such as the T7 promoter for prokaryotes or CMV for eukaryotic cells.
5. Observe Reporter Genes
If the plasmid contains reporter genes, such as fluorescent proteins (e.g., EGFP) or enzyme markers (e.g., β-galactosidase), assess their expression to quickly determine the success of foreign gene expression.
6. Validate Primer Binding Sites
Confirm the locations and sequences of primer binding sites to facilitate primer design for PCR amplification or sequencing validation.
CD Genomics offers sequencing services to generate comprehensive plasmid maps and uncover various plasmid-related information, including plasmid genes and other essential features.
What is the Function of Plasmids?
Plasmids, as small, independent DNA molecules, play significant roles across various disciplines, including genetic engineering, molecular biology, and vaccine development. Their ability to autonomously replicate, carry foreign genes, and facilitate gene transfer between cells positions them as essential tools in modern biotechnology.
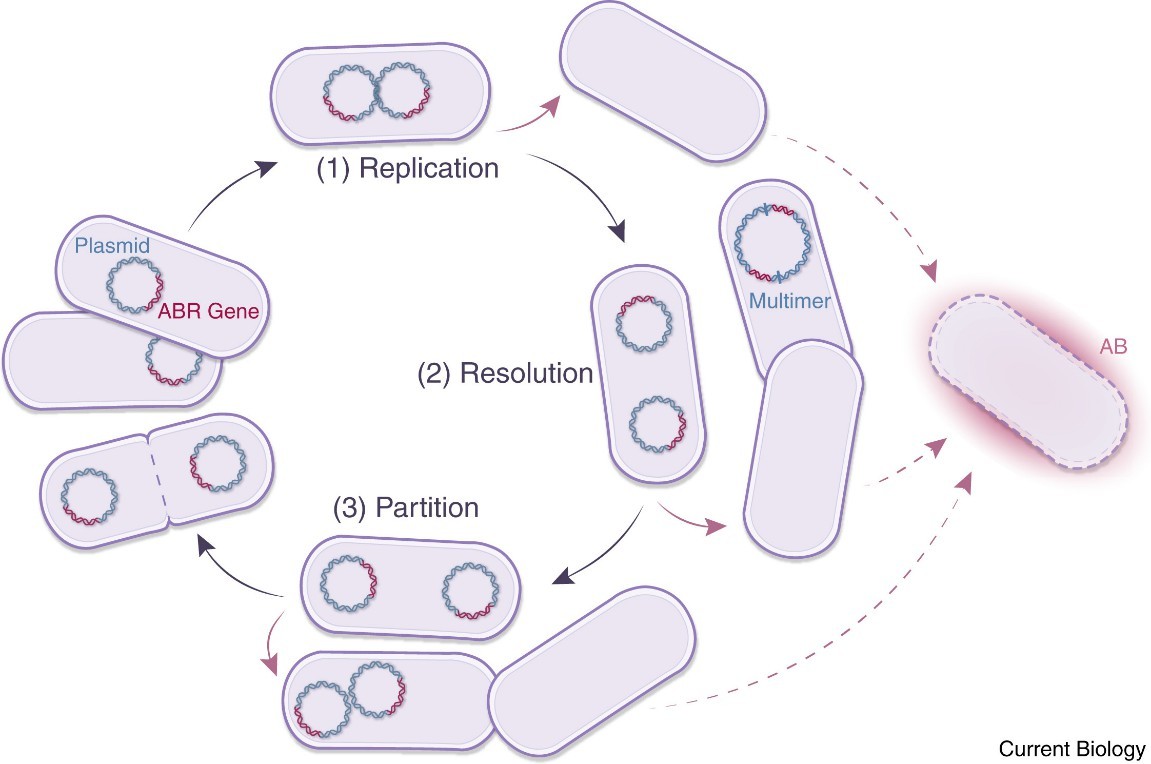 Plasmid life cycle. (Wein, et al., Current Biology, 2020)
Plasmid life cycle. (Wein, et al., Current Biology, 2020)
Common Types of Plasmids and Their Functions
- Cloning Plasmids: These plasmids are utilized for the replication and amplification of foreign DNA fragments within host cells. Typically, they contain multiple restriction enzyme sites that facilitate the insertion of target genes and include antibiotic resistance genes serving as selectable markers.
- Expression Plasmids: Designed to express specific proteins within host cells, expression plasmids include essential elements such as promoters, resistance genes, and terminators, enabling the regulation of transcription and translation of target genes.
- Shuttle Plasmids: These versatile plasmids can transfer between different host cells, including bacteria, yeast, and mammalian cells. They usually incorporate replication origins and selective markers specific to particular hosts.
- Integrative Plasmids: These are engineered to integrate foreign genes into the host cell's genome. Such plasmids contain homologous recombination sequences that allow precise pairing and integration with the target genomic DNA.
- Reporter Plasmids: Containing reporter genes like green fluorescent protein (GFP) or β-galactosidase, these plasmids are employed to monitor gene expression, protein localization, or cellular signaling pathways.
- Silencing Plasmids: Used in RNA interference (RNAi) experiments, silencing plasmids express small interfering RNA (siRNA) or short hairpin RNA (shRNA) to downregulate specific gene expression.
- CRISPR/Cas Plasmids: These plasmids contain components of the CRISPR/Cas system, utilized for precise gene editing by targeting specific locations in the genome for cleavage, facilitating gene knockout or insertion.
- Resistance Plasmids: Harboring resistance genes, these plasmids are critical for selecting cells that can withstand specific antibiotics, proving particularly useful during cloning to identify successfully transformed cells.
- Artificial Chromosome Plasmids: Larger in design, these plasmids can accommodate bigger DNA fragments, making them especially useful in genomic research and gene therapy applications.
The significance of plasmids extends beyond laboratory research. In natural settings, they play a pivotal role in horizontal gene transfer among bacteria, enabling rapid acquisition of new traits, such as antibiotic resistance. This capability is essential for understanding bacterial evolution and microbial ecology. In industrial and agricultural biotechnology, plasmids are harnessed for the production of recombinant proteins, enhancement of crop traits, and the development of novel therapies.
What is the Difference Between a DNA Vector and a Plasmid?
In the fields of genetic engineering and molecular biology, both vectors and plasmids serve as critical tools for introducing exogenous DNA into host cells for applications such as gene cloning, expression, and research. However, these two concepts differ significantly in terms of definition, functionality, and application. A nuanced understanding of these distinctions is essential for scientific research and technological applications.
Concept of Vectors
In genetic engineering, a vector is defined as a DNA molecule that facilitates the transfer of exogenous genes into recipient cells. Vectors act as vehicles that transport foreign genes from one organism to another or within cellular environments. Within molecular biology experiments, the primary function of a vector is to introduce a target gene into host cells while ensuring that these genes can replicate or express within the host.
Essential Characteristics and Functions of Vectors:
- Efficient Gene Transfer: Vectors can successfully deliver exogenous genes into recipient cells, facilitating gene transfer.
- Replication or Integration Capability: Vectors provide necessary replication origins or integration sites to ensure that the genes can be amplified or integrated within host cells.
- Selection Markers: Vectors typically contain specific selection markers (e.g., antibiotic resistance genes) to facilitate the identification and selection of successfully transformed cells.
- Manipulability: Vectors must possess multiple cloning sites to accept the insertion of exogenous DNA and suitable restriction sites for gene cloning purposes.
Common types of vectors include:
- Plasmid Vectors
- Viral Vectors
- Phage Vectors
Primary Differences Between Vectors and Plasmids
Although plasmids are among the most commonly used vectors in genetic engineering, notable differences exist between vectors and plasmids concerning their definitions and applications:
| Comparison Dimension |
Vectors |
Plasmids |
| Definition |
Broadly defined DNA molecules capable of carrying exogenous genes into host cells. This category includes plasmids, viruses, and phages, depending on experimental requirements. |
Naturally occurring, circular DNA molecules that exist independently of host chromosomes, commonly found in bacteria and eukaryotic cells. They represent a subset of vectors. |
| Function |
Introduce exogenous genes, provide replication or integration capabilities, and must possess selection markers and restriction sites for gene manipulation. |
Autonomous replication and stable inheritance; used in gene cloning, protein expression, gene therapy, and intercellular gene transfer. |
| Application Range |
Broadly applicable in gene transfer, gene therapy, vaccine development, etc. Viral vectors target mammalian cells, while phage vectors are specific to bacteria. |
Primarily employed in bacterial genetic engineering (e.g., gene cloning, expression, and recombinant protein production). Plasmids may also be used for gene transfection in yeast and other eukaryotic organisms. |
| Structure |
Structural diversity exists, with vectors manifesting as linear or circular DNA/RNA (viral vectors) or as viral particle structures (phage vectors). |
Typically circular DNA with functional elements such as replication origins (Ori), antibiotic resistance genes, selection markers, promoters, and cloning sites. |
| Applicable Cells |
Selection of target host cells is dependent on vector type, e.g., viral vectors for mammalian cells and phage vectors for bacteria. |
Primarily utilized in prokaryotic cells such as bacteria or yeast, with limited applicability in higher eukaryotic cells. |
Understanding these differences enhances our grasp of genetic manipulation techniques and underscores the integral roles that both vectors and plasmids play in contemporary biotechnology.
If you're interested in learning more about plasmids and their sequencing, consider reading:
Plasmid Detection and Complete Plasmid DNA Sequencing
Plasmid Fact Sheet: Definition, Structure and Application
Exploring Plasmid Extraction: Techniques and Key Considerations
Mastering Whole Plasmid Sequencing: Key Insights and Benefits
References:
- Helinski, Donald R. "A brief history of plasmids." EcoSal Plus 10.1 (2022): eESP-0028. https://doi.org/10.1128/ecosalplus.ESP-0028-2021
- Wein, Tanita, and Tal Dagan. "Plasmid evolution." Current Biology 30.19 (2020): R1158-R1163. DOI: 10.1016/j.cub.2020.07.003
- Zhao, Baisong, et al. "Kindlin-1 regulates astrocyte activation and pain sensitivity in rats with neuropathic pain." Regional Anesthesia & Pain Medicine 43.5 (2018): 547-553. https://doi.org/10.1097/AAP.0000000000000780
- Tao, Y., Zhou, K., Xie, L. et al. Emerging coexistence of three PMQR genes on a multiple resistance plasmid with a new surrounding genetic structure of qnrS2 in E. coli in China. Antimicrob Resist Infect Control 9, 52 (2020). https://doi.org/10.1186/s13756-020-00711-y
- Smalla, Kornelia, Sven Jechalke, and Eva M. Top. "Plasmid detection, characterization, and ecology." Microbiology spectrum 3.1 (2015): 10-1128. https://doi.org/10.1128/microbiolspec.plas-0038-2014
- Garcillán-Barcia, Maria Pilar, Andres Alvarado, and Fernando de la Cruz. "Identification of bacterial plasmids based on mobility and plasmid population biology." FEMS microbiology reviews 35.5 (2011): 936-956. https://doi.org/10.1111/j.1574-6976.2011.00291.x


 Sample Submission Guidelines
Sample Submission Guidelines

 Analysis results of integrated ATAC-seq and RNA-seq results.
Analysis results of integrated ATAC-seq and RNA-seq results. Plasmid life cycle. (Wein, et al., Current Biology, 2020)
Plasmid life cycle. (Wein, et al., Current Biology, 2020)