CD Genomics provides complete plasmid and phage sequencing services. Our advanced bioinformatics pipelines support de novo assembly, eliminating the need for reference genomes and enabling thorough and precise analysis of both plasmids and bacteriophages.
What is Complete Plasmid/Phage Sequencing
Plasmids are circular DNA molecules that exist independently within bacterial cells, often carrying genes that endow their host with beneficial traits such as antibiotic resistance or enhanced metabolic functions. These plasmids are essential for horizontal gene transfer, allowing bacteria to swiftly adapt to environmental pressures and change. Similarly, phages, or bacteriophages, are viruses that specifically target bacteria and play a crucial role in bacterial gene transfer and population dynamics. Their interactions with bacterial hosts impact microbial diversity and ecological balance.
A thorough understanding of the genetic structure of plasmids and phages is vital for grasping their roles in microbial ecosystems and their influence on traits like antibiotic resistance. Complete Plasmid/Phage Sequencing involves decoding the entire genetic sequence of plasmids and phages, offering an extensive view of their genetic landscape. This method allows researchers to identify both the essential genes and the accessory elements that contribute to the organism's adaptability and pathogenic potential. Through this detailed genetic mapping, we gain deeper insights into microbial evolution and gene transfer processes, which are crucial for advancing research in biotechnology and environmental science.
Our Whole Plasmid/Phage Sequencing Methods
Plasmids and phages exhibit considerable variability in size and complexity, with plasmids often showing high GC content, intricate structures, or extensive repetitive regions, while phages can also display diverse genetic architectures and variable genome sizes. These complexities have historically made traditional Sanger sequencing methods both expensive and prone to high failure rates, coupled with relatively slow processing times.
| Aspect | Sanger Sequencing | Whole Plasmid/Phage Sequencing (NGS) |
|---|---|---|
| Methodology | Chain-termination method | Utilizes Next-Generation Sequencing (NGS) or Long Read Sequencing technologies |
| Sequence Length | Suitable for shorter fragments (up to 1,000 bp) | Capable of sequencing entire plasmids and phages, regardless of size or complexity |
| Speed | Sanger Sequencing can involve extended processing times and is particularly suited for small-scale projects. | Whole Plasmid/Phage Sequencing using NGS technologies offers a faster turnaround, ideal for high-throughput applications |
| Cost | Can be cost-prohibitive for larger plasmids due to multiple sequencing reactions | Generally more cost-effective, especially for larger plasmids and phages, as it requires fewer sequencing reactions |
| Accuracy | Sanger Sequencing is renowned for its high precision and minimal error rates | Whole Plasmid/Phage Sequencing maintains exceptional accuracy and incorporates advanced error correction methods |
| Applicability | Sanger Sequencing is effective for smaller plasmids but may face challenges with larger, intricate, or repetitive structures | Whole Plasmid/Phage Sequencing is highly versatile, accommodating a wide range of plasmid and phage sizes and complexities, making it suitable for comprehensive genomic analysis |
CD Genomics employs advanced sequencing technologies to deliver complete plasmid and phage DNA sequences. Our methods integrate both next-generation sequencing (NGS) and long-read sequencing technologies to accommodate the diverse structures and sizes of plasmids and phages. We utilize state-of-the-art platforms such as Illumina for high-throughput sequencing and Nanopore or PacBio for long-read sequencing. Our approach includes comprehensive library preparation and bioinformatics analysis, ensuring accurate assembly and annotation of the entire genetic sequence.
Advantages of Complete Plasmid/Phage Sequencing
Comprehensive Coverage: Provides full-length sequencing of plasmids and phages, including regions with high GC content and intricate structures that traditional methods might miss, ensuring a complete genetic overview.
High Accuracy: Delivers sequences with accuracy exceeding 99.9%, ensuring the genetic information is precise and dependable for further research.
Cost-Effective: Offers a more affordable solution, especially for large plasmids and phages, with costs significantly lower compared to traditional sequencing methods, making it a cost-efficient choice for extensive projects.
Rapid Turnaround: Ensures quick processing and data acquisition, accelerating research and enabling timely insights.
No Reference Required: Performs sequencing without the need for pre-existing reference genomes, ideal for exploring novel or uncharacterized plasmids and phages.
High Throughput: Capable of handling multiple samples simultaneously, enhancing efficiency and supporting large-scale research efforts.
Applications of Complete Plasmid/Phage Sequencing
- Microbial Ecology: Examine the role of plasmids and phages within microbial communities, their interactions, and their influence on microbial diversity and ecosystem functions.
- Gene Transfer Studies: Investigate horizontal gene transfer mechanisms mediated by plasmids and phages, including the dissemination of resistance genes and other functional attributes.
- Synthetic Biology: Utilize comprehensive plasmid sequences to design and refine genetic constructs, advancing applications in synthetic biology.
- Phage Research: Analyze phage genomes to develop strategies for targeted interventions against specific bacterial species.
- Environmental Research: Explore the presence and functions of plasmids and phages across diverse environments, such as soil, water, and various microbial habitats.
Complete Plasmid/ Phage Sequencing Workflow

Service Specifications
| Sample Requirements Complete Plasmid DNA Sequencing:
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
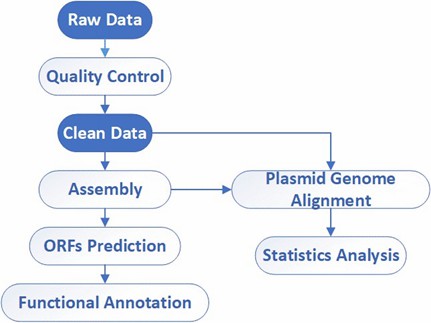
Deliverables
- FASTQ files
- Read-length and quality report
- Assembly results
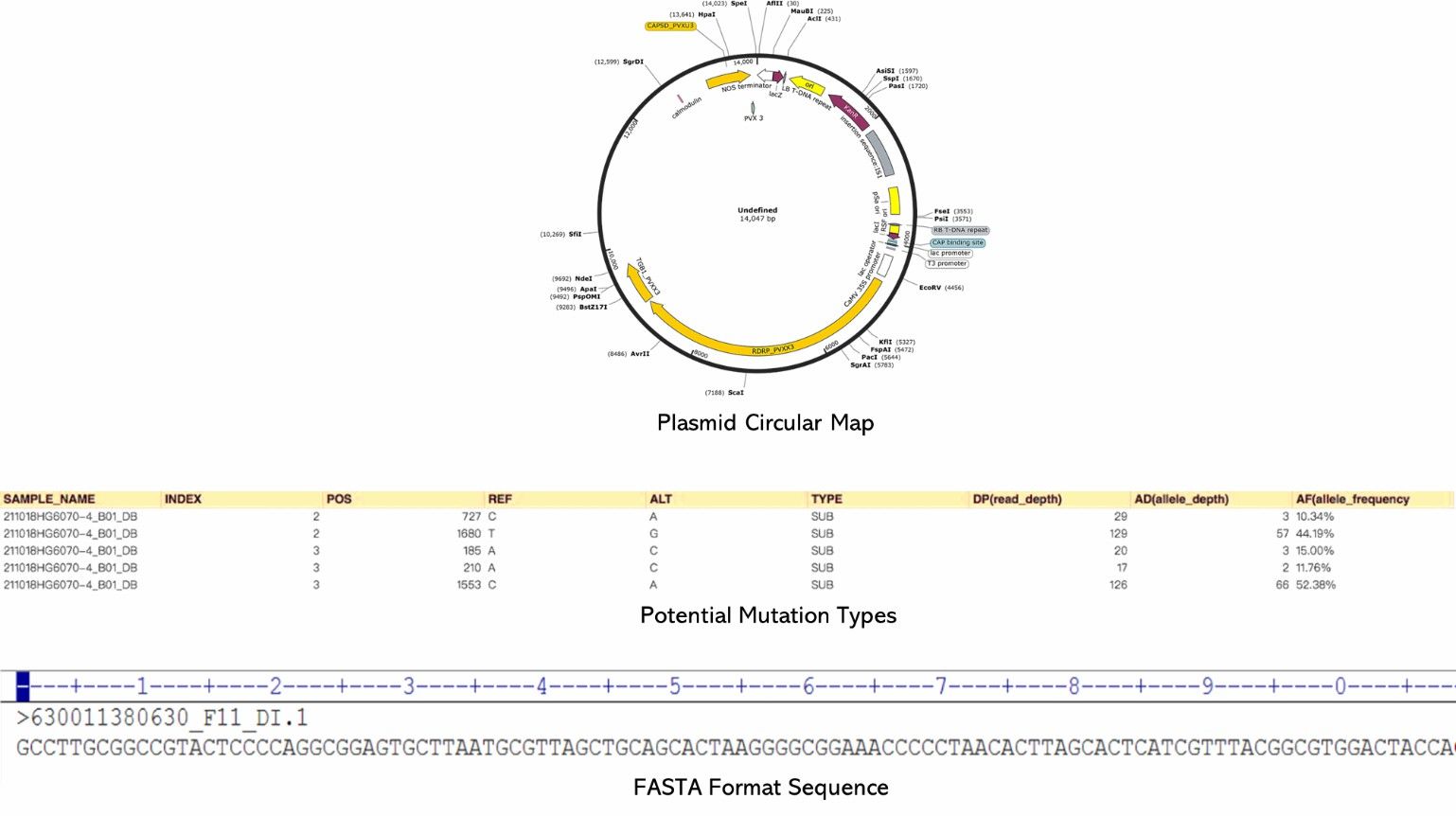
- Genomic circular map
- Analysis of potential mutations
- Annotation files
CD Genomics leverages the advanced capabilities of the Illumina Platform for precise plasmid sequencing, ensuring reliable verification and comprehensive analysis of plasmid DNA. In addition, we have developed a cost-effective, high-throughput method using Nanopore technology to efficiently sequence larger plasmids in their entirety. Extending our expertise to phage sequencing, we utilize these cutting-edge technologies to provide thorough and accurate genomic insights into bacteriophages. With our extensive experience and state-of-the-art platforms, we are dedicated to delivering exceptional sequencing services and high-quality results tailored to meet the diverse needs of our clients.
1. Why is complete plasmid DNA sequencing necessary?
- Validating sequence integrity: Plasmids may undergo genetic modifications during cloning or amplification, introducing potential errors or mutations.
- Quality control for cloning and engineering: Sequencing plasmids ensures accurate verification of cloned inserts, absence of mutations, and experimental system integrity.
- Optimizing experimental design: Accurate plasmid sequences enable effective experimental planning.
2. How to extract plasmid DNA from host cells?
Common extraction methods include:
- Alkaline lysis: Cell lysis under alkaline conditions to release plasmid DNA.
- Phenol-chloroform extraction: Utilizing phenol and chloroform for DNA extraction, eliminating proteins and impurities.
- Column-based extraction: Commercial columns for plasmid DNA purification through centrifugation and elution.
3. How to ensure sequencing data accuracy?
Critical measures to ensure the precision of sequencing data encompass several key procedures:
- Thorough sample extraction: Guaranteeing the purity and concentration of plasmid DNA.
- Selecting a suitable sequencing platform: Tailoring the choice to experimental demands.
- Implementing data processing and quality control: Employing bioinformatics tools to eliminate substandard reads and sequencing errors.
- Validating through sequence alignment: Aligning the results with known sequences to substantiate accuracy.
4. What are the applications of complete plasmid DNA sequencing?
Complete plasmid DNA sequencing boasts a plethora of applications across various domains:
- Gene cloning and expression studies: Facilitating the construction of effective cloning vectors.
- Antibiotic resistance research: Unraveling bacterial resistance mechanisms.
- Gene function studies: Delving into specific gene functionalities within cells.
- Synthetic biology and biotechnology: Pioneering the design of synthetic biology tools and pathways for pharmaceutical production, environmental restoration, and bioenergy ventures.
If you would like to learn more, please refer to our article "Plasmid Detection and Complete Plasmid DNA Sequencing."
5. What factors may affect the results of plasmid DNA sequencing?
Influencing factors encompass:
- Sample quality: DNA purity and concentration adequacy.
- Sequencing technology: Appropriateness of chosen platforms and technologies.
- Data processing: Effectiveness of quality control and processing procedures.
- Sequence complexity: Impact of plasmid sequence complexity and repetitive elements on assembly.
Dynamics of Antimicrobial Resistance and Genomic Epidemiology of Multidrug-Resistant Salmonella enterica Serovar Indiana ST17 from 2006 to 2017 in China
Journal: Msystems
Impact factor: 7.324
Published: 21 July 2022
Background
Nontyphoidal Salmonella enterica (NTS) is a significant global foodborne pathogen, increasingly associated with antimicrobial resistance (AMR), especially multidrug-resistant (MDR) strains. This resistance complicates treatment and poses a public health challenge, leading the WHO to prioritize resistant S. enterica for new antimicrobial development. The genetic basis of resistance includes chromosomal mutations and plasmid-mediated genes, contributing to the widespread dominance of certain strains. Recent studies in China highlight the prevalence and resistance mechanisms of MDR S. Indiana, underscoring the urgent need for effective interventions to curb its spread.
Methods
- 251 S. Indiana isolates
- Fecal samples
- Clinical samples and food-related samples
- Antimicrobial susceptibility testing
- DNA extraction
- Whole-genome sequencing
- Detection of AMR genotypes
- Analysis of blaCTX-M genomic locations
- Phylogenetic analysis
Results
Among 138 blaCTX-M-positive isolates, 65.2% were classified by genome location. Notably, blaCTX-M-14 and blaCTX-M-55 were identified on chromosomes, while blaCTX-M-15 and blaCTX-M-65 were predominantly carried by plasmids. Human isolates exhibited a significantly higher prevalence of blaCTX-M positivity (63%) compared to food-related isolates (47%). Complete sequencing of five representative isolates unveiled a diverse landscape of genetic contexts harboring blaCTX-M genes, particularly on IncHI2 plasmids. Remarkably, plasmid pIndS104-CTX displayed striking similarity to a chromosomal locus in S. Indiana SI43, implying potential recombination events between plasmids and chromosomes. This study underscores the complexity of blaCTX-M dissemination pathways among pathogens and emphasizes the urgent need for further elucidation.
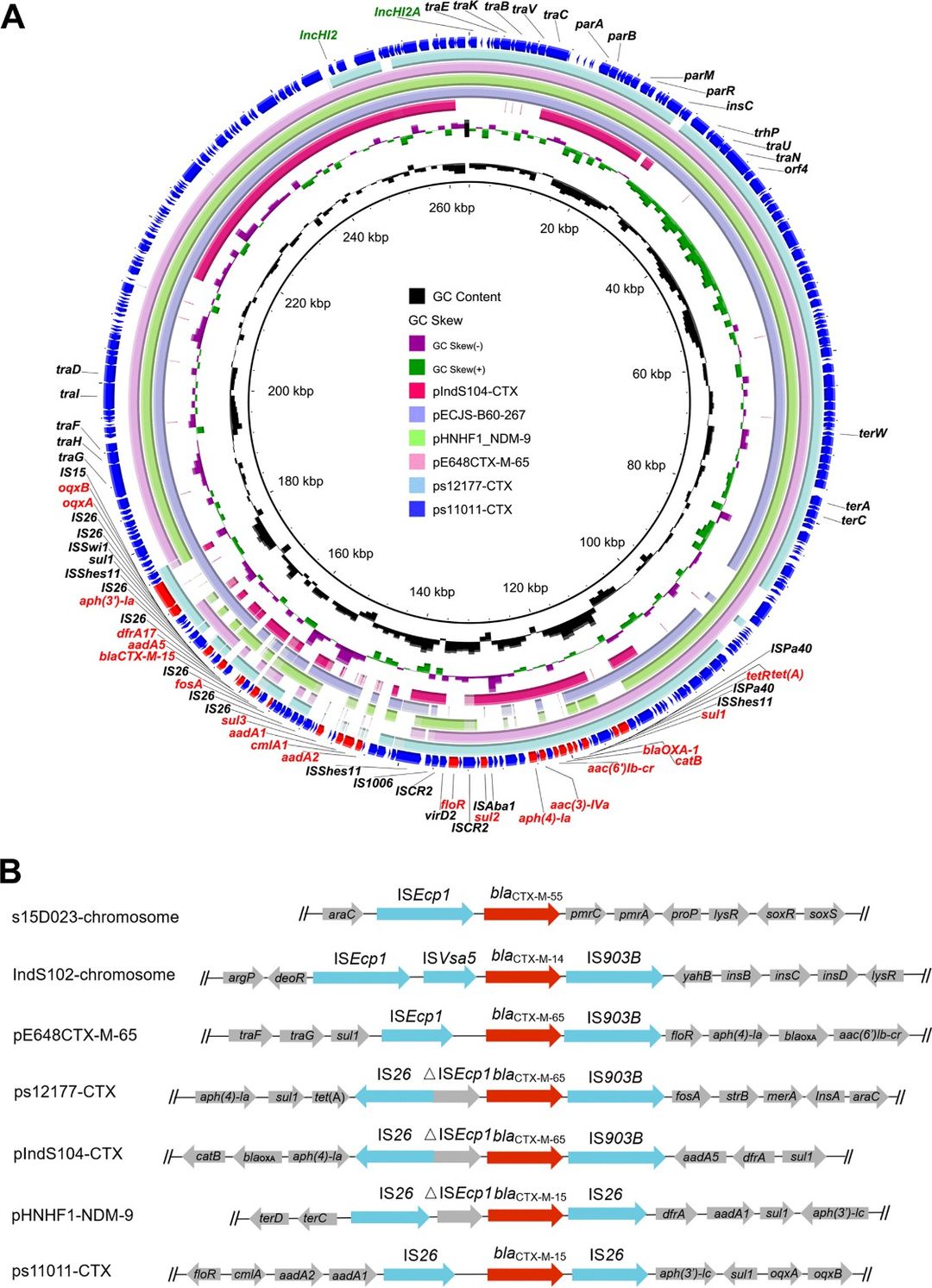 Fig 1. Circular comparison between blaCTX-M-positive IncHI2 plasmids in this study (pIndS104-CTX, ps17177-CTX, and ps11011-CTX) and other similar IncHI2 plasmids in the NCBI nr database.
Fig 1. Circular comparison between blaCTX-M-positive IncHI2 plasmids in this study (pIndS104-CTX, ps17177-CTX, and ps11011-CTX) and other similar IncHI2 plasmids in the NCBI nr database.
This nationwide study of 251 isolates found strong genomic similarity between human and chicken isolates, suggesting chickens as a source of human infections. Lineage 6 was the most resistant, with IncHI2 plasmids commonly carrying ESBL and PMQR genes.
Conclusion
This study offers a detailed insight into the rapid evolution of multidrug resistance (MDR) in S. Indiana over the past 15 years in China. Unique antimicrobial resistance mechanisms distinguish S. Indiana from other serovars, with diverse genetic processes contributing to resistance development, including chromosomal integrations, evolution of mobile resistance elements, and sporadic acquisition of resistance determinants. The presence of diverse host niches, including various animal reservoirs, underscores the importance of a One Health approach for efficient monitoring and control of resistance spread. Continuous surveillance targeting bacterial strains and mobile genetic elements is essential for effective control measures.
Reference:
- Du P, Liu X, Liu Y, et al. Dynamics of antimicrobial resistance and genomic epidemiology of multidrug-resistant Salmonella enterica serovar Indiana ST17 from 2006 to 2017 in China. Msystems, 2022, 7(4): e00253-22.
Here are some publications that have been successfully published using our services or other related services:
Distinct functions of wild-type and R273H mutant Δ133p53α differentially regulate glioblastoma aggressiveness and therapy-induced senescence
Journal: Cell Death & Disease
Year: 2024
High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing
Journal: International Journal of Molecular Sciences
Year: 2020
Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI
Journal: New phytologist
Year: 2017
Generation of a highly attenuated strain of Pseudomonas aeruginosa for commercial production of alginate
Journal: Microbial Biotechnology
Year: 2019
Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Journal: Viruses
Year: 2024
Genome Analysis and Replication Studies of the African Green Monkey Simian Foamy Virus Serotype 3 Strain FV2014
Journal: Viruses
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
