Shotgun metagenomics represents a transformative advancement in environmental science, enabling researchers to analyze microbial ecosystems in unprecedented detail. By extracting and directly sequencing genetic material from environmental specimens, this technique eliminates traditional requirements for microbial cultivation or DNA amplification. The comprehensive genomic data obtained reveals not only the taxonomic diversity of microorganisms present but also their metabolic capabilities and potential functions. This powerful approach has become instrumental in uncovering complex ecological relationships, characterizing previously unknown species, and understanding the biochemical cycles that drive environmental processes.
Definition of Shotgun Metagenomics
The fundamental principle of shotgun metagenomics involves extracting DNA from environmental specimens and performing non-targeted sequencing of the fragmented genetic material. Through this comprehensive analytical approach, researchers can elucidate both the species composition and metabolic capabilities of diverse microorganisms within a given ecosystem. The resulting genomic data encompasses information about numerous microbial groups, including bacterial species, archaeal populations, and viral entities. In contrast to conventional laboratory cultivation techniques, this sequencing-based methodology provides a more complete understanding of microbial ecosystems, particularly revealing insights about organisms that resist traditional growth methods.
Techniques and Tools in Shotgun Metagenomic Sequencing
Common sequencing platforms
Shotgun Metagenomic Sequencing is a high-throughput sequencing technology widely used in microbial community research. The core of this is to randomly interrupt, sequence and assemble all genomic DNA in a sample to comprehensively assess microbial diversity and its function. Currently, commonly used sequencing platforms include Illumina, Oxford Nanopore and PacBio.
Illumina: This platform is known for its short read lengths (150-300 bp) and high-throughput output, making it suitable for genome assembly and annotation. For example, Illumina's HiSeq series of platforms are commonly used for microbial metagenome sequencing and can provide output of up to 150 Gb with high accuracy and consistency.
Oxford Nanopore: This platform is characterized by long read lengths (1-100 kb) and real-time sequencing capabilities, making it particularly suitable for solving assembly problems of complex genomic regions. However, its error rate is high, so it is often necessary to combine short read long technology to improve accuracy.
PacBio: This platform is also known for its long read lengths (1-10 kb), but its low error rate is suitable for high-quality genome assembly. PacBio technology has also been widely used in microbial metagenome research.
Service you may interested in
Resource
Data analysis tools and bioinformatics workflows
Data analysis for bullet metagenomic sequencing involves multiple steps, including quality control, data processing, assembly and annotation. The following are the main bioinformatics tools and workflows:
Quality control: Quality control is the first step in data analysis. Common tools include Trimmomatic, Ktrim, Cutadapt and MultiQC. These tools are used to remove low-quality reads, splice sequences and other noise.
Data processing: The data processing stage includes comparing reference databases (such as the NCBI database) or building de novo assemblies. Commonly used assembly software include MegaHit, MetaVelvet, MetaSPAdes, etc.
Annotations and functional analysis: After assembly, the genome needs to be functionally annotated to identify the functional genes and metabolic pathways of the microorganism. For example, the KEGG database can be used to analyze microbial metabolic pathways, while TaxonFinder can be used to classify species.
Advantages of Shotgun Metagenomic Sequencing in Environmental Studies
Comprehensive Diversity Analysis
Bullet-style metagenomic sequencing allows comprehensive sequencing of the genomes of all microorganisms in a sample, including bacteria, archaea, viruses and other types of microorganisms. This method avoids the limitations of traditional culture methods and can reveal the diversity of rare and uncultured microorganisms, thereby providing more comprehensive information on the composition of microorganisms. For example, by analyzing Sargasso Sea samples, researchers found more than 1200 unknown bacterial genotypes, as well as a large number of photoreceptor-related genes.
Functionality Insights
Compared with traditional methods based on target gene or functional screening, bullet-based metagenomic sequencing can directly analyze the genomic information of microbial communities and infer their functional potential. For example, the technology could reveal the role of microorganisms in ecosystem processes such as nitrogen cycling, carbon fixation, and antibiotic resistance. In addition, it can also be used to identify potential pathogens and antibiotic resistance genes, providing important clues for disease diagnosis and treatment.
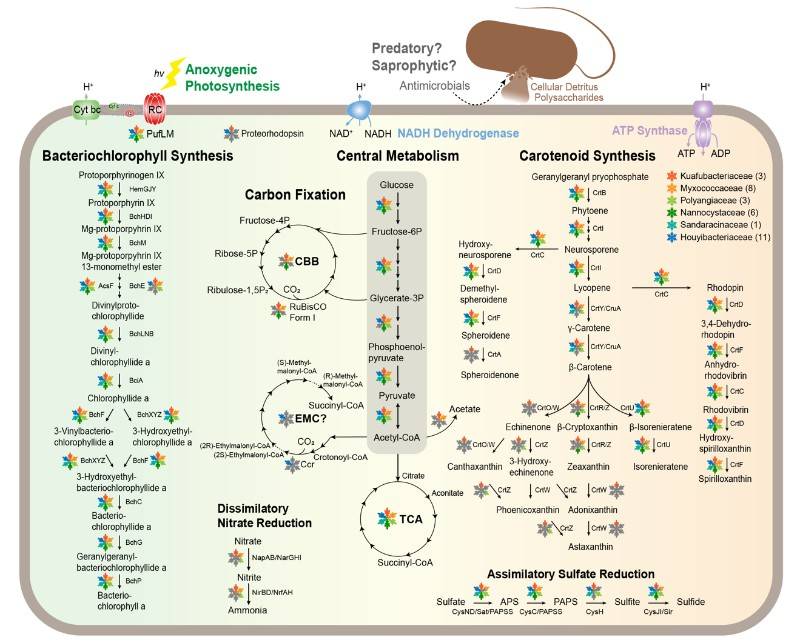 Fig. 1. Metabolic schemes of potential phototrophic Myxococcota.(Li,et.al,2023)
Fig. 1. Metabolic schemes of potential phototrophic Myxococcota.(Li,et.al,2023)
Resolving Complex Ecosystems
Shotgun metagenomics sequencing is particularly suitable for the analysis of complex microbial ecosystems, such as soil, water and air samples. It can simultaneously detect genomic information from a large number of microorganisms, helping researchers understand the interactions between microbial communities and their responses to environmental factors such as temperature, pH and nutrients.
High resolution
Compared with other metagenomic technologies, bullet sequencing has higher resolution and can distinguish between species and subspecies level differences. This high resolution allows it to more accurately assess the composition and functional potential of microbial communities. For example, through long-read and long-sequencing technology, researchers can reconstruct a more complete microbial genome and better interpret its functional characteristics.
Case Studies in Environmental Shotgun Metagenomics
Environmental Shotgun Metagenomics is a powerful technology that studies the composition, function and interaction with the environment of microbial communities by extracting and sequencing DNA directly from environmental samples without the need to cultivate microorganisms. The following is a detailed analysis of three case studies.
Case Study 1: Soil microbiome analysis to understand soil health and fertility.
Soil health and fertility are critical for sustainable agriculture, and the soil microbiome plays an essential role in these processes. The analysis of soil microbiomes can provide insights into the interactions between microbial communities and soil health, which can ultimately influence crop yield and quality.
A research conducted in Brazil explored the relationship between soil fertility levels and microbial community diversity in tropical pastures. The study found that high-fertility soils exhibited greater bacterial diversity, particularly among nitrogen-cycling groups, which are crucial for maintaining pasture productivity. This illustrates how understanding the soil microbiome can inform management practices that enhance soil fertility and agricultural sustainability.
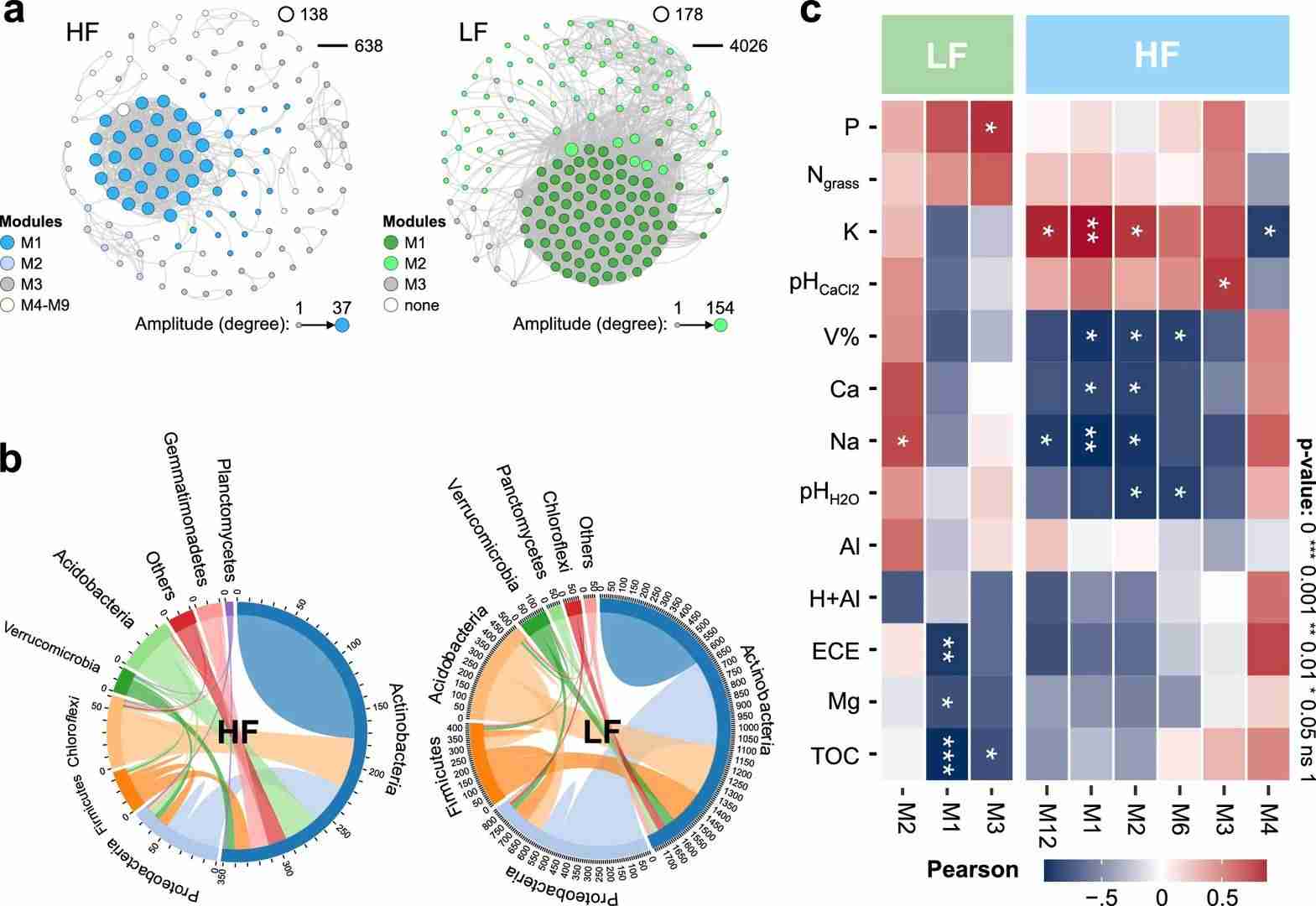 Fig. 2. Microbial co-occurrence analyses in pastures on fertile (HF) and poor (LF) soil.(Paes da Costa,et.al,2024)
Fig. 2. Microbial co-occurrence analyses in pastures on fertile (HF) and poor (LF) soil.(Paes da Costa,et.al,2024)
A study examined the impact of a seven-species multi-mix cover crop treatment on soil nutrient content and microbial diversity. The results indicated that multi-mix cover cropping significantly enhanced microbial abundance and diversity compared to single-species cover crops. This practice not only improved soil health but also contributed to higher crop yields by promoting beneficial microbial communities that aid in nutrient cycling and plant growth(Wang,et.al,2020).
Case Study 2: Monitoring water quality and microbial contaminants in aquatic ecosystems.
Monitoring water quality is essential for maintaining healthy aquatic ecosystems, as it helps identify microbial contaminants that can pose risks to both environmental and human health. Effective monitoring programs assess various parameters, including physical, chemical, and biological indicators, to ensure the integrity of water bodies.
In practical applications, microorganisms play an important role in water remediation and pollution control. For example, certain microorganisms can degrade organic pollutants, thereby improving water quality. At the same time, microorganisms can also serve as "sentries" of water ecosystems, reflecting the health status of water bodies by monitoring changes in their number and distribution. For example, in the study of Lake Victoria, shotgun method metagenomic analysis revealed the diversity of microbial communities and its relationship with pollution, which provided a new perspective for understanding pollution mechanisms(Khatiebi,et.al, 2023).
Case Study 3: Identifying pathogens in environmental air samples using shotgun metagenomics.
The use of shotgun metagenomics for identifying pathogens in environmental air samples has emerged as a powerful tool for understanding microbial diversity and potential health risks associated with airborne pathogens. This approach allows for comprehensive analysis of complex microbial communities without the need for prior culturing, making it particularly valuable in monitoring environments such as hospitals, schools, and urban areas.
An innovative study utilized nanopore sequencing-based metagenomics to assess the air microbiome in urban environments like Barcelona. By actively sampling air and analyzing the microbial composition, researchers were able to identify diverse microbial taxa and their potential implications for human health. This approach not only provided insights into the urban air quality but also highlighted the need for ongoing surveillance to track emerging pathogens that could impact public health.
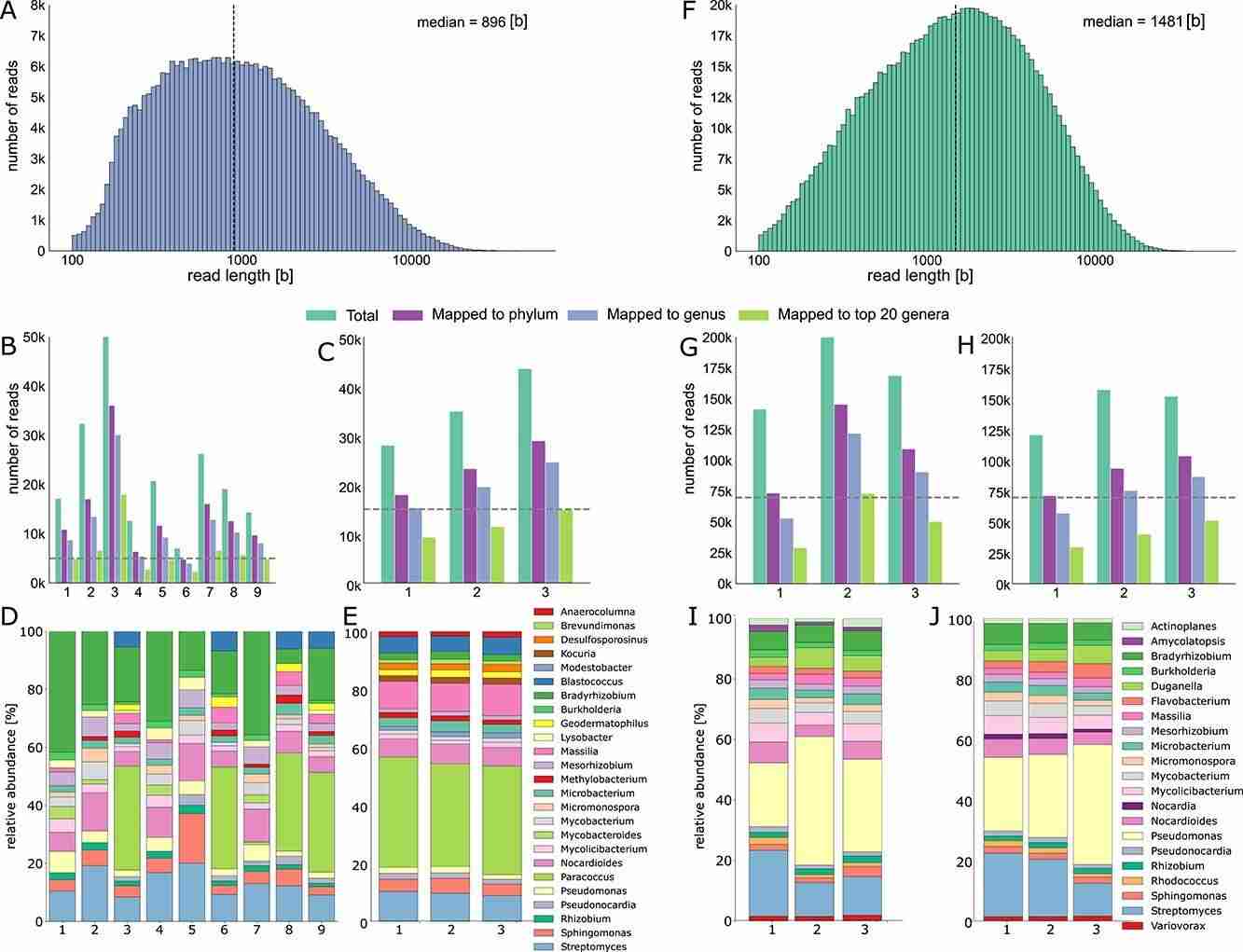 Fig. 3. Robust air microbiome assessments of a controlled and natural environment .(Reska,et.al,2024)
Fig. 3. Robust air microbiome assessments of a controlled and natural environment .(Reska,et.al,2024)
In conclusion, shotgun metagenomics is a transformative tool in environmental studies, offering unparalleled insight into microbial diversity, functionality, and ecosystem dynamics. By enabling the study of uncultured microorganisms and their roles in critical ecological processes, this technology paves the way for a deeper understanding of the natural world.
References:
- Li, L., Huang, D., Hu, Y., et.al. (2023). Globally distributed Myxococcota with photosynthesis gene clusters illuminate the origin and evolution of a potentially chimeric lifestyle. Nature communications, 14(1), 6450. https://doi.org/10.1038/s41467-023-42193-7
- Paes da Costa, D., das Graças Espíndola da Silva, T.,et.al. (2024). Soil fertility impact on recruitment and diversity of the soil microbiome in sub-humid tropical pastures in Northeastern Brazil. Scientific reports, 14(1), 3919. https://doi.org/10.1038/s41598-024-54221-7
- Wang, C. H., Wu, L., Wang, Z., Alabady, M. S., Parson, D., Molumo, Z., & Fankhauser, S. C. (2020). Characterizing changes in soil microbiome abundance and diversity due to different cover crop techniques. PloS one, 15(5), e0232453. https://doi.org/10.1371/journal.pone.0232453
- Khatiebi, S., Kiprotich, K., Onyando, Z., Wekesa, C., Chi, C. N., Mulambalah, C., & Okoth, P. (2023). Shotgun Metagenomic Analyses of Microbial Assemblages in the Aquatic Ecosystem of Winam Gulf of Lake Victoria, Kenya Reveals Multiclass Pollution. BioMed research international, 2023, 3724531. https://doi.org/10.1155/2023/3724531
- Reska, T., Pozdniakova, S., Borràs, S.,et.al. (2024). Air monitoring by nanopore sequencing. ISME communications, 4(1), ycae099. https://doi.org/10.1093/ismeco/ycae099


 Sample Submission Guidelines
Sample Submission Guidelines
 Fig. 1. Metabolic schemes of potential phototrophic Myxococcota.(Li,et.al,2023)
Fig. 1. Metabolic schemes of potential phototrophic Myxococcota.(Li,et.al,2023) Fig. 2. Microbial co-occurrence analyses in pastures on fertile (HF) and poor (LF) soil.(Paes da Costa,et.al,2024)
Fig. 2. Microbial co-occurrence analyses in pastures on fertile (HF) and poor (LF) soil.(Paes da Costa,et.al,2024) Fig. 3. Robust air microbiome assessments of a controlled and natural environment .(Reska,et.al,2024)
Fig. 3. Robust air microbiome assessments of a controlled and natural environment .(Reska,et.al,2024)