The nitrogen cycle stands as one of Earth's paramount biogeochemical cycles, with microorganisms playing an indispensable role in its intricate web. Harnessing the power of metagenomic technologies, we unlock a comprehensive view of both the sheer quantity and the diverse array of functional microbial taxa interwoven within the nitrogen cycle. Consequently, we gain profound insights into the underlying patterns and intricate mechanisms governing this pivotal cycle.
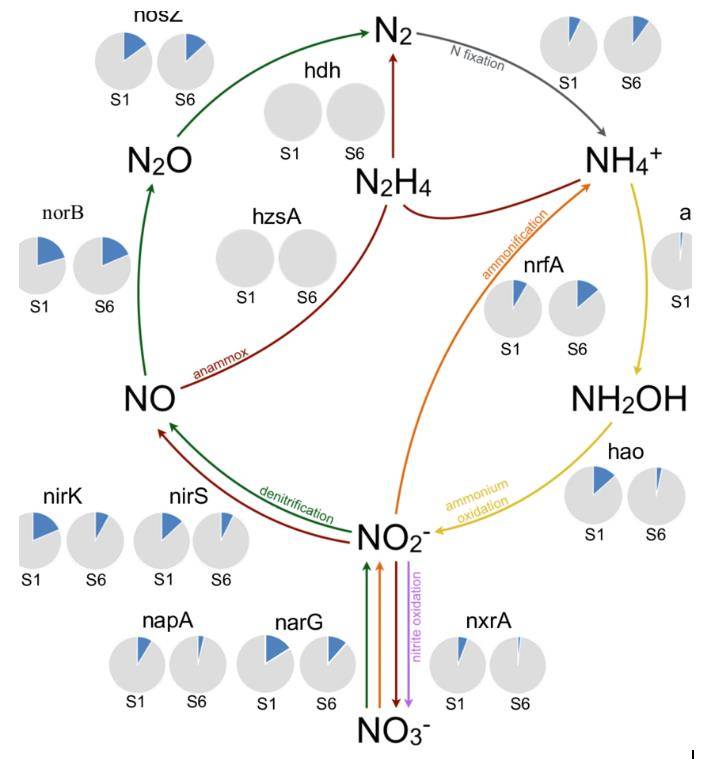 Overview of nitrogen cycle processes by selected marker genes. (de Jong et al., 2018)
Overview of nitrogen cycle processes by selected marker genes. (de Jong et al., 2018)
Nitrogen Cycle
Nitrogen, a life-sustaining element, holds the distinction of being a chief limiting factor for biological growth on our planet. While atmospheric nitrogen (N2) abounds, most organisms depend on bioavailable nitrogen forms, such as ammonium and nitrate, to fuel their growth. The availability of these nitrogen forms hinges upon a series of intricate nitrogen transformation reactions, collectively referred to as the nitrogen cycle. This cycle orchestrates the movement, alteration, and perpetual recycling of elemental nitrogen between the Earth's atmosphere, biosphere, soil, and hydrosphere.
The nitrogen cycle can be dissected into six distinct nitrogen transformation processes, each serving a unique purpose:
- Assimilation: Organisms convert inorganic nitrogen forms (e.g., ammonium salts and nitrates) into organic nitrogen, a vital resource for growth and synthesis in living entities.
- Ammonification: The breakdown of organic nitrogen compounds into ammonium salts marks the conversion from organic to inorganic nitrogen.
- Nitrification: A conversion process wherein ammonium salts metamorphose into nitrates, featuring two sub-processes: ammonia oxidation and nitrite oxidation.
- Denitrification: Nitrate undergoes reduction to elemental nitrogen (N2), liberating nitrogen into the atmosphere.
- Anaerobic Ammonia Oxidation (Anammox): A specialized nitrogen transformation pathway that directly converts ammonium and nitrate into nitrogen gas, bypassing intermediates.
- Nitrogen Fixation: Atmospheric nitrogen (N2) undergoes conversion into biologically accessible nitrogen forms, such as ammonium salts.
The fluxes between these nitrogen transformation processes exhibit considerable variation, with the conversion between ammonia and organic nitrogen being one of the highest flux processes, while nitrification stands as a significant flux contributor.
Metagenomics in Nitrogen Cycle Research
Metagenomic technologies have emerged as indispensable tools in unraveling the mysteries of microorganisms and their associated genes within the nitrogen cycle. The following highlights key facets and databases integral to metagenomic studies focused on the nitrogen cycle:
- Identification of Nitrogen Cycle-Associated Genes: Metagenomic data mining facilitates the discovery of nitrogen cycle-associated genes residing in the natural environment. These genes encode enzymes and proteins crucial for nitrogen transformation.
- Quantification of Gene Abundance: Metagenomic data empowers us to gauge the relative abundance of nitrogen cycle-related genes among diverse microbial groups, thereby illuminating their significance in ecosystems.
- Species Distribution of Associated Genes: Metagenomic data provides insights into the distribution of nitrogen cycle-related genes across various microbial species, shedding light on the role of microbial diversity in shaping the nitrogen cycle's intricate tapestry.
Nitrogen Cycle Databases
Databases collectively provide a rich source of genetic information related to the nitrogen cycle, allowing researchers to delve into the intricacies of nitrogen metabolism and its associated genes. The specialized databases (NcycFunGen and NCycDB) offer comprehensive and user-friendly resources, while KEGG provides additional context but may require more intricate navigation, especially for incomplete gene data. Researchers can choose the most suitable database(s) based on their specific research needs and the completeness of their gene sequences.
NcycFunGen Database
This invaluable database houses a substantial collection of genetic data, encompassing 60,735 amino acid sequences, nucleic acid sequences, and HMM profiles of 22 pivotal genes associated with the nitrogen cycle.
Researchers can harness these resources for a multitude of purposes. Amino acid sequences can be organized into mmseqs sequence libraries, facilitating systematic indexing. Homology matching can then be applied for detailed annotation. Furthermore, HMM spectra can be employed for spectral sequence searches and comprehensive annotation of Open Reading Frame (ORF) sequences.
The database also offers a mapping of amino acid sequences to their respective gene symbols, streamlining the exploration of nitrogen cycle genes.
NCycDB Database
The NCycDB database is a treasure trove comprising 273,942 amino acid sequences, which are further refined to 273,501 by eliminating duplicate sequence identifiers. These sequences correspond to a total of 68 unique genes associated with the nitrogen cycle.
Researchers can tap into this resource for a broad spectrum of applications. The amino acid sequence database provides a comprehensive foundation for genomic investigations. Additionally, the database offers a valuable mapping that associates amino acid sequences with their corresponding gene symbols.
KEGG Database
KEGG, a renowned bioinformatics resource, features an array of genes related to the nitrogen cycle, which are linked to KEGG KOs (KEGG Orthology).
KEGG's extensive data can be used to access KEGG KO HMM spectra, offering insights into gene functions. Moreover, it provides access to amino acid and nucleic acid sequences associated with KEGG genes.
While KEGG is a valuable resource, it's important to note that it may yield fewer annotation results compared to specialized databases like NcycFunGen and NCycDB. Particularly, the KEGG kofamscan method might prove less user-friendly, especially for incomplete gene sequences.
Case: Studying Nitrogen Cycling in Cold Seep Environments through Metagenomic Sequencing
The Enigma of Diazotrophs in Cold Seep Sediments
Recent research had identified anaerobic methanotrophic archaea (ANME-2) and their sulfate-reducing bacterial partners (SEEP-SRB1 clade) as diazotrophic pioneers in deep-sea cold seep sediments. These remarkable organisms, through their nitrogen-fixing abilities, challenge the harsh conditions of the cold seep environment. Yet, a pressing question loomed: Are ANME-2 and SEEP-SRB1 the sole nitrogen-fixing champions in this icy abyss?
The Quest for Comprehensive Insights
To unveil the hidden dimensions of nitrogen cycling in cold seeps, this intrepid team embarked on a quest. Armed with cutting-edge metagenomic sequencing techniques, they collected and analyzed data from 61 metagenomes, 1428 metagenome-assembled genomes, and six metatranscriptomes drawn from 11 cold seep locations worldwide. Their mission: to decipher the intricate web of nitrogen cycling in these mysterious depths.
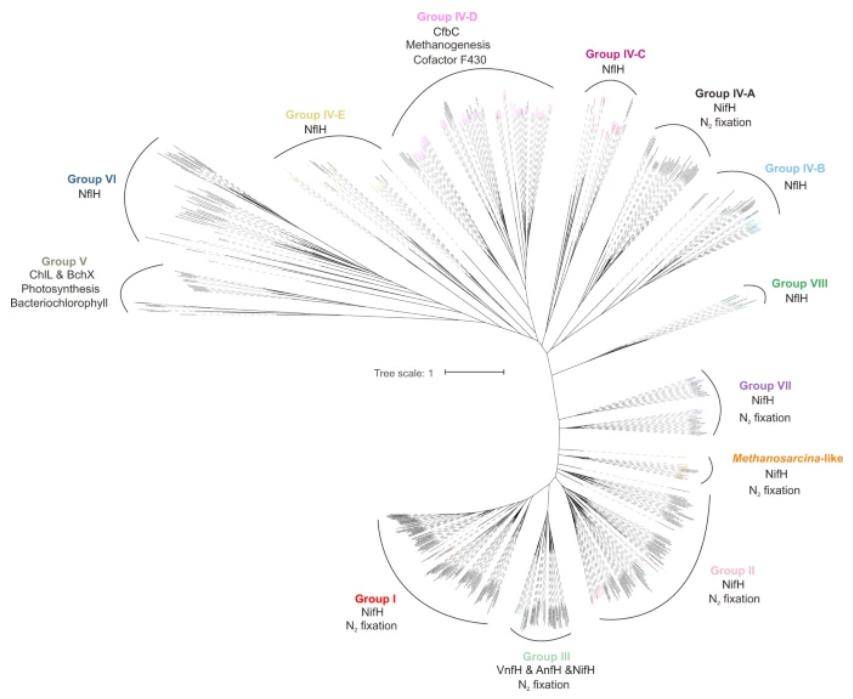 Maximum-likelihood phylogenetic tree of non-redundant nitrogenase subunit NifH identified from cold seep metagenomic assemblies. (Dong et al., 2022)
Maximum-likelihood phylogenetic tree of non-redundant nitrogenase subunit NifH identified from cold seep metagenomic assemblies. (Dong et al., 2022)
Revealing the Diversity of Diazotrophic Lineages
The metagenomic journey unveiled a remarkable revelation—a diverse array of nitrogenase genes spanning an expansive diversity of diazotrophic lineages nestled within cold seep sediments. The nitrogen-fixing prowess, it appeared, was not confined solely to ANME-2 and SEEP-SRB1. Instead, a rich tapestry of microbial groups emerged as potential nitrogen fixers, rewriting the narrative of nitrogen cycling in these extreme environments.
Unlocking the Mechanisms
The quest didn't stop at gene discovery; they delved deeper into the metabolic machinery driving nitrogen fixation. Predictions of diverse catabolic pathways hinted at the means by which these cold seep microorganisms powered their nitrogen-fixing abilities, challenging the prevailing notion that diazotrophy in these environments was intrinsically linked to sulfate-dependent anaerobic oxidation of methane.
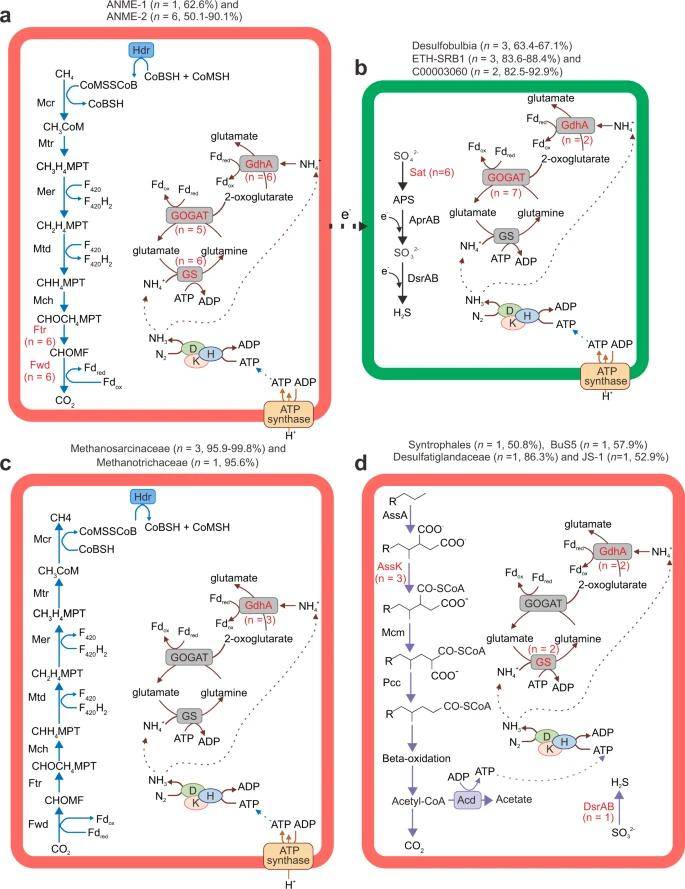 Metabolic reconstruction of core pathways for nitrogen-fixing MAGs. (Dong et al., 2022)
Metabolic reconstruction of core pathways for nitrogen-fixing MAGs. (Dong et al., 2022)
Genetic Mobility and Evolutionary Forces
Intriguingly, evidence of genetic mobility among nitrogen fixation genes within the diazotrophic groups in cold seeps was uncovered. Moreover, the forces of evolution, in the form of purifying selection, seemed to shape these genetic traits. This suggested a dynamic and adaptable nitrogen-fixing community in the cold seep ecosystem.
Expanding the Horizon
Their findings pushed the boundaries of nitrogen cycling research in cold seep environments. Five candidate phyla, Altarchaeia, Omnitrophota, FCPU426, Caldatribacteriota, and UBA6262, emerged as potential diazotrophs, expanding the roster of nitrogen-fixing organisms in these remote and harsh habitats. These previously overlooked nitrogen fixers might, they postulated, be substantial contributors to the global nitrogen balance.
References:
- Dong, Xiyang, et al. "Phylogenetically and catabolically diverse diazotrophs reside in deep-sea cold seep sediments." Nature Communications 13.1 (2022): 4885.
- de Jong, Anniek, et al. "Decrease in microbial diversity along a pollution gradient in Citarum River Sediment." BioRxiv (2018): 357111.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Overview of nitrogen cycle processes by selected marker genes. (de Jong et al., 2018)
Overview of nitrogen cycle processes by selected marker genes. (de Jong et al., 2018) Maximum-likelihood phylogenetic tree of non-redundant nitrogenase subunit NifH identified from cold seep metagenomic assemblies. (Dong et al., 2022)
Maximum-likelihood phylogenetic tree of non-redundant nitrogenase subunit NifH identified from cold seep metagenomic assemblies. (Dong et al., 2022) Metabolic reconstruction of core pathways for nitrogen-fixing MAGs. (Dong et al., 2022)
Metabolic reconstruction of core pathways for nitrogen-fixing MAGs. (Dong et al., 2022)