In recent years, the cycle time for second-generation sequencing library construction has seen remarkable improvements. The second-generation short-read sequencing technology continues to maintain a dominant position in the sequencing market. However, since its inception in 2008, third-generation sequencing technology has been advancing at an impressive rate. With its unique advantage of long-read capabilities and PCR-free sequencing process, it has enabled the individual sequencing of each DNA molecule. This technology is now widely applied in various areas such as genome assembly, pathogen research, and mutation identification.
In the realm of genomics, Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have emerged as the leading pioneers in long-read sequencing. They are actively pushing the boundaries of complex genome analysis, structural variation detection, and real-time biological research. PacBio's Single Molecule Real-Time (SMRT) sequencing technology and Nanopore's nanopore-based electrical signal detection capitalize on their unique principles to overcome the limitations of short-read technologies. These have become essential tools in precision medicine, pathogen monitoring, and evolutionary biology research.
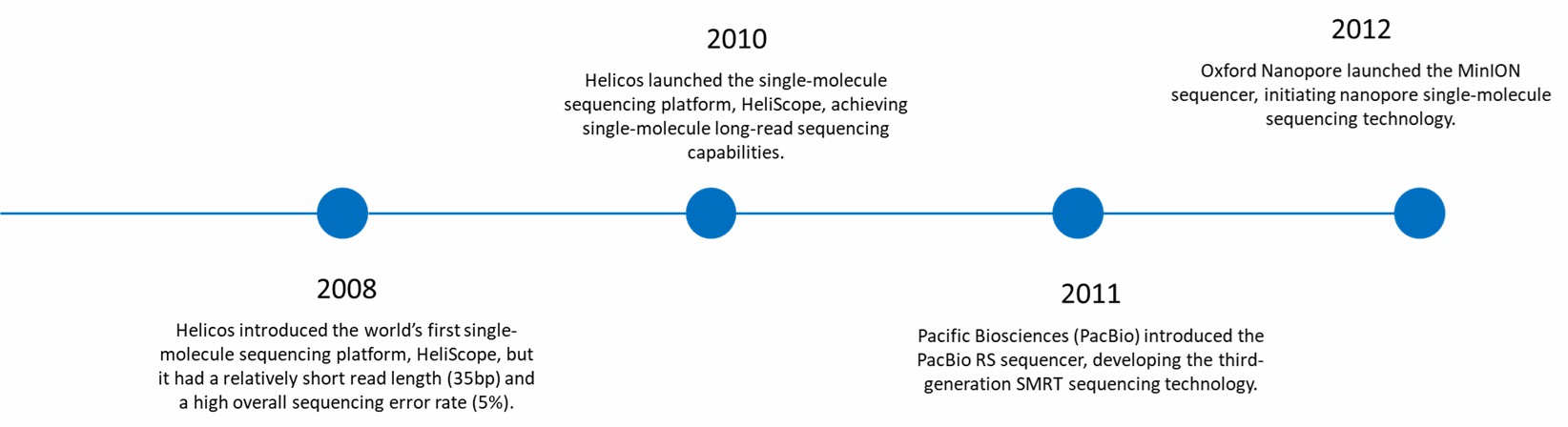 Development of Long-read Sequencing Technology (2008-2012).
Development of Long-read Sequencing Technology (2008-2012).
Comparison of PacBio and Nanopore Technologies
The core differences between PacBio and Nanopore technologies are reflected in sequencing principles, read length and accuracy, as well as throughput and cost. PacBio employs SMRT sequencing technology to record DNA synthesis through fluorescent signals, generating highly accurate HiFi reads. In contrast, Nanopore sequencing detects DNA sequences using nanopore electrical current, enabling real-time data streaming and ultra-long reads. In terms of throughput and cost, PacBio's Sequel IIe is well-suited for large-scale, high-precision projects, while Nanopore's PromethION is renowned for its flexibility and lower entry cost. Below, a detailed comparative analysis is offered from dimensions of principles, read length and accuracy, and throughput and cost, along with a summarized tabular representation.
Principle Differences: SMRT Sequencing (Enzyme-Driven) vs. Nanopore Electrical Signal (Electrochemical Detection)
PacBio's SMRT technology leverages zero-mode waveguides (ZMW) and DNA polymerase, where fluorescently labeled dNTPs are excited by a laser to record the base synthesis in real-time. Its primary advantage lies in the production of high-fidelity HiFi reads, achieving single-molecule accuracy levels exceeding >99.9% through cyclic consensus sequencing (CCS) to correct random errors.
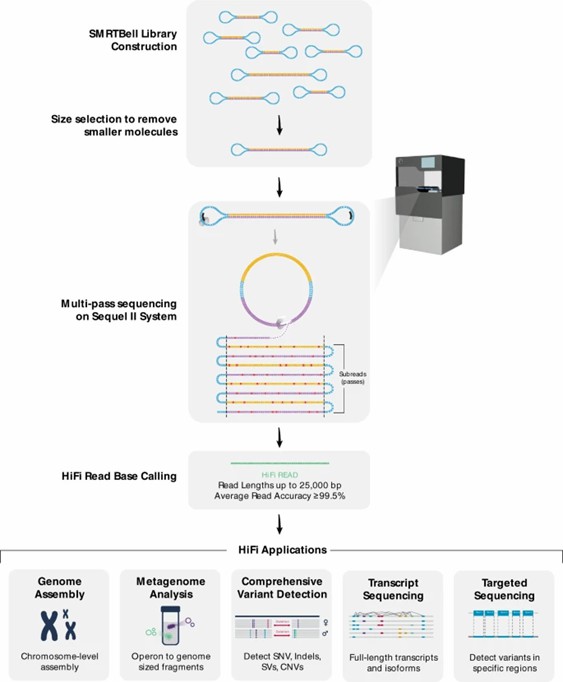 Flowchart of HiFi sequence read generation and downstream applications. (Hon, T., et al., 2020)
Flowchart of HiFi sequence read generation and downstream applications. (Hon, T., et al., 2020)
Conversely, Nanopore technology operates on electrochemical detection principles, where single-stranded DNA traversing a protein nanopore induces specific current changes, directly translating into sequence information. Nanopore technology requires no amplification or labeling, supporting real-time data flow and direct detection of epigenetic modifications such as methylation. The latest R10 chip, with its dual-reader head design, significantly enhances accuracy in homopolymeric regions.
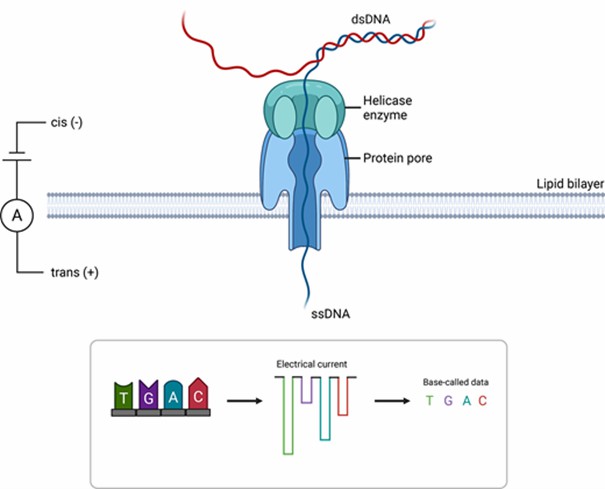 A schematic diagram of the mechanism of Oxford Nanopore Technologies (ONT) sequencing. (Beckett, Angela H., et al., 2021)
A schematic diagram of the mechanism of Oxford Nanopore Technologies (ONT) sequencing. (Beckett, Angela H., et al., 2021)
Read Length and Accuracy: PacBio's High-Fidelity Model vs. Nanopore's Real-Time Sequencing Characteristics
PacBio's Sequel IIe platform achieves an average enzyme read length exceeding 70 kb, with HiFi reads reaching 10–20 kb, and post-error correction accuracy surpassing 99.9%, ideal for high-accuracy assembly and structural variation detection. Nanopore's PromethION provides single-molecule read lengths up to megabase levels, with an N50 of approximately 35 kb. The R10 chip's initial read accuracy has been improved to 93.8%, and consensus sequences (at 50X coverage) reach Q44 (99.996%).
Throughput and Cost: Comparative Platform Throughput (e.g., PromethION vs. Sequel IIe)
PacBio's Sequel IIe generates 120 Gb of HiFi data per run, fitting medium to large-scale projects, though it entails higher equipment costs and complex sample preparation. Nanopore's PromethION offers a throughput of up to 1.9 Tb per run, supports flexible scaling, and features lightweight equipment (e.g., MinION), catering to budget-constrained or field-based applications.
Technology Comparison Summary Table
| Comparison Dimension |
PacBio |
Nanopore |
| Principle |
Fluorescently labeled dNTPs + ZMW |
Nanopore current sensing |
| Read Length |
10–20 kb (HiFi) |
Up to Mb levels |
| Initial Accuracy |
~85% |
~93.8% (R10 chip) |
| Corrected Accuracy |
>99.9% |
~99.996% (consensus sequence, 50X depth) |
| Throughput |
120 Gb/run (HiFi) |
1.9 Tb/run (PromethION) |
| Equipment Cost |
High |
Low (MinION portable) |
| Suitable Applications |
Clinical research, genome assembly |
Field monitoring, real-time analysis |
Through this comparative analysis and the summary table, it is evident that PacBio excels in accuracy and the detection of epigenetic modifications, whereas Nanopore stands out in real-time capability, portability, and ultra-long reads. The choice of technology should be carefully balanced against research objectives (such as assembly integrity and real-time requirements) and budget considerations.
Application Scenarios of PacBio and ONT
PacBio and Nanopore each offer unique advantages across various research domains, with their specific technological characteristics rendering them indispensable in certain settings. PacBio excels in structural variation analysis and transcriptome research due to its high-precision HiFi reads and capacity for epigenetic modification detection. On the other hand, Nanopore's real-time capability and portability provide unmatched utility in field monitoring and on-the-spot clinical diagnostics. Below is a detailed analysis of their applications in specific scenarios.
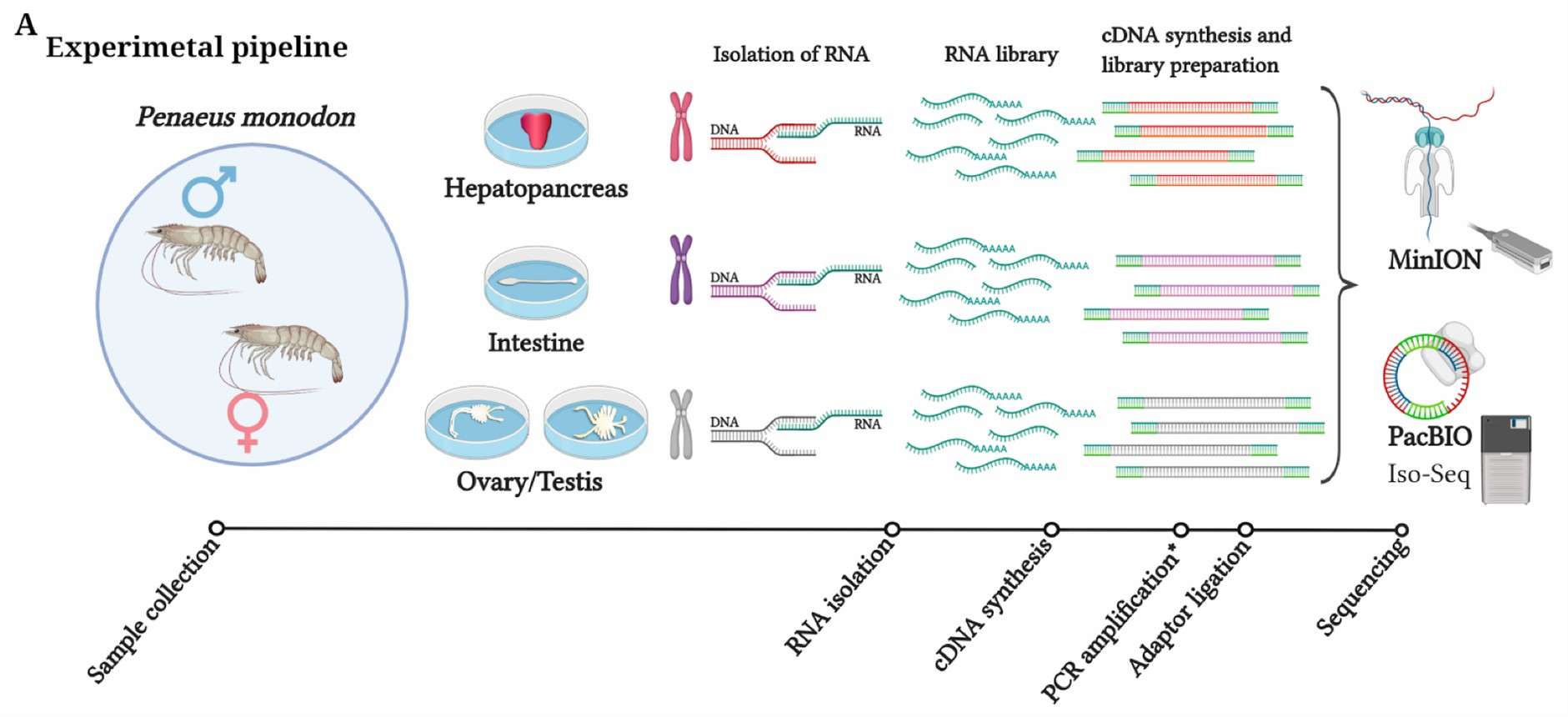 Experimental pipelines used in this study. Asterisk (*) indicates a step in the pipeline required for PacBio sequencing. (Udaondo, Zulema, et al. 2021)
Experimental pipelines used in this study. Asterisk (*) indicates a step in the pipeline required for PacBio sequencing. (Udaondo, Zulema, et al. 2021)
PacBio: Detection of Structural Variations and Precision Transcriptome Analysis
PacBio's HiFi reads technology stands out in the analysis of complex genomes, particularly beneficial in the following applications:
- Structural Variation Detection: HiFi reads accurately identify structural variations such as insertions, deletions, inversions, and translocations, supplying critical data for cancer genomics and rare disease research.
- Precision Transcriptome Analysis: HiFi reads enable full-length transcript sequencing, capturing RNA isoforms directly and revealing gene expression regulatory mechanisms.
- Epigenetic Research: PacBio can directly detect DNA methylation, such as 5mC, and other base modifications, providing high-resolution data for epigenetic regulatory network studies.
Nanopore: Real-Time Monitoring and Portability in Various Scenarios
Nanopore's real-time sequencing capacity and portability make it highly advantageous for monitoring dynamic biological processes and in-field applications:
- Real-Time Pathogen Monitoring: Nanopore supports real-time data streaming, allowing for pathogen genome sequencing and analysis within hours, suitable for rapid response to infectious disease outbreaks.
Portable Sequencing Scenarios: The lightweight and portable MinION device is suited for genome sequencing in field, polar, and even space environments.
- Clinical Point-Of-Care Testing (POCT): Nanopore technology enables bedside testing for rapid pathogen diagnostics or patient genome variant analysis.
- Direct RNA Sequencing: Nanopore technology reads RNA sequences and modifications directly without reverse transcription, offering new perspectives for functional genomics research.
Application Scenario Comparison Table
| Technology |
Core Application |
Unique Value |
Typical Case Studies |
| PacBio |
Structural Variation Detection |
High-precision decoding of complex genomes |
Cancer genomics, rare disease research |
|
Precision Transcriptome Analysis |
Reconstruction of full-length isoforms |
Discovery of plant alternative splicing events |
|
Epigenetic Research |
Direct detection of DNA methylation |
Tumor-specific methylation pattern analysis |
| Nanopore |
Real-Time Pathogen Monitoring |
Rapid infectious disease outbreak response |
Ebola virus field monitoring |
|
Portable Sequencing Scenarios |
Genome sequencing in extreme conditions |
Microgravity environment sequencing on ISS |
|
Clinical Point-Of-Care Testing |
Rapid bedside diagnostics |
Quick detection of genetic diseases in newborn ICUs |
|
Direct RNA Sequencing |
RNA modification and dynamic analysis |
Full-length genome research of RNA viruses |
Through the comparative analysis and table, it is clear that while PacBio demonstrates superior performance in accuracy and epigenetic modification detection, Nanopore excels in real-time functionality, portability, and ultra-long read capabilities. The choice of technology should be strategically aligned with research objectives, such as assembly integrity and real-time needs, as well as budget constraints.
Services you may interested in
Learn More
Summary of Advantages and Limitations
PacBio and Nanopore technologies each possess distinctive advantages and limitations. Choosing between them necessitates careful consideration of research objectives, budgetary constraints, and data requirements. Below is a comprehensive summary of the strengths and weaknesses of each technology in terms of accuracy, flexibility, cost, and data processing.
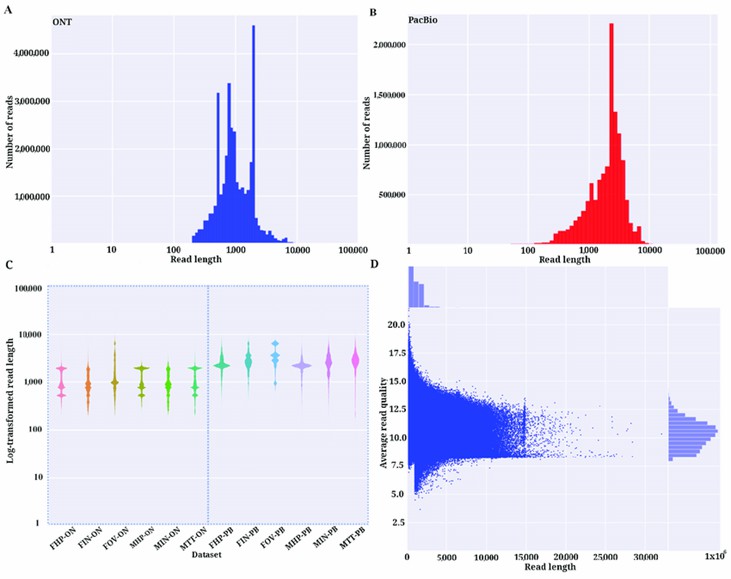 Comparison of ONT and PacBio sequencing libraries. (Udaondo, Zulema, et al., 2021)
Comparison of ONT and PacBio sequencing libraries. (Udaondo, Zulema, et al., 2021)
Advantages of PacBio
- High Accuracy: PacBio's HiFi reads, utilizing cyclic consensus sequencing (CCS) to correct random errors, achieve post-correction accuracy rates exceeding 99.9%. This makes it ideal for high-precision genome assembly and variation detection.
- Epigenetic Modification Detection: Capable of detecting DNA methylation (such as 5mC) and base modifications directly without additional experimentation, offering high-resolution data for epigenetic studies.
- Long-Read Capability: With enzyme read lengths averaging over 70 kb and HiFi reads reaching 10–20 kb, PacBio can effectively resolve complex repetitive regions and structural variations.
Limitations of PacBio
- High Equipment Cost: Initial investment in the PacBio Sequel IIe platform is substantial, making it more suitable for laboratories with ample budgets.
- Complex Sample Preparation: Library preparation involves intricate procedures and requires high-quality DNA samples.
- Limited Throughput: While HiFi data quality is high, the per-run throughput is relatively low, best suited for medium to small-scale projects.
Advantages of Nanopore
- Real-Time Sequencing: Supports real-time data streaming, allowing for immediate results during sequencing, which is beneficial in time-sensitive scenarios like infectious disease monitoring.
- Portability: The MinION device is lightweight and portable, facilitating genomic sequencing in field, polar, and even space environments.
- Ultra-Long Reads: Capable of single-molecule read lengths up to megabase levels, enabling it to span complex genomic regions and reduce assembly complexity.
- Lower Cost: Compared to PacBio, Nanopore's lower equipment costs make it accessible for research teams with limited budgets.
Limitations of Nanopore
- Higher Initial Error Rate: Despite advancements with the R10 chip and consensus sequence techniques that significantly enhance accuracy, the raw read error rate remains higher than PacBio's HiFi reads.
- Reliance on Post-Processing: Data analysis requires sophisticated error-correction algorithms and bioinformatics tools, increasing the complexity of analysis.
- Sample Requirements: Even though library preparation is relatively straightforward, high standards for DNA integrity and purity are necessary.
Comparison Table of Advantages and Limitations
| Technology |
Advantages |
Limitations |
| PacBio |
- High accuracy (HiFi mode) |
- High equipment cost |
|
- Epigenetic modification detection |
- Complex sample preparation |
|
- Long-read capability |
- Limited throughput |
| Nanopore |
- Real-time sequencing |
- Higher initial error rate |
|
- Portability |
- Reliance on post-processing optimization |
|
- Ultra-long read capability |
- High sample requirements |
|
- Low cost |
|
In conclusion, the choice between PacBio and Nanopore technologies depends on specific research needs, including the desired balance between accuracy and throughput, the necessity for portability, and financial considerations. Making this decision requires a nuanced understanding of each platform's capabilities and constraints.
PacBio vs Nanopore: Selection Guidelines
When considering whether to utilize PacBio or Nanopore technology, it is essential to weigh research objectives, budget constraints, and specific data requirements. The following recommendations provide detailed guidance across dimensions such as genome assembly, real-time capability, cost, and application scenarios, aiding researchers in selecting the most appropriate technology.
1. Technology Selection Based on Research Objectives
| Research Objective |
Recommended Technology |
Justification |
| High-Precision Genome Assembly |
PacBio |
HiFi reads offer post-error correction accuracy >99.9%, making them ideal for assembling complex genomes and detecting structural variations. |
| Real-Time Monitoring and Rapid Response |
Nanopore |
Supports real-time data streaming, suitable for infectious disease monitoring and timely clinical diagnostics. |
| Full-Length Transcriptome Analysis |
PacBio |
HiFi reads facilitate full-length transcript sequencing, directly capturing RNA isoforms and elucidating gene expression regulatory mechanisms. |
| Sequencing in Extreme Environments |
Nanopore |
The portable MinION device is well-suited for genomic sequencing in field, polar, and even space environments. |
| Epigenetic Research |
PacBio |
Direct detection of DNA methylation and base modifications provides high-resolution data for studying epigenetic regulatory networks. |
| Large-Scale Pathogen Genome Research |
Nanopore |
With a PromethION platform throughput of up to 1.9 Tb per run, it supports high-throughput pathogen genome sequencing and analysis. |
2. Technology Selection Based on Budget
| Budget Range |
Recommended Technology |
Justification |
| High Budget (>$500,000) |
PacBio |
The Sequel IIe platform requires a significant initial investment, but its superior HiFi data quality is suitable for high-precision research projects. |
| Moderate Budget ($100,000-$500,000) |
Nanopore |
The PromethION platform offers moderate costs and high throughput, aligning with the needs of medium-sized laboratories. |
| Low Budget (<$100,000) |
Nanopore |
The low-cost MinION device is ideal for budget-constrained research teams or field applications. |
3. Technology Selection Based on Data Needs
| Data Requirement |
Recommended Technology |
Justification |
| High-Precision Data |
PacBio |
HiFi reads, with post-error correction accuracy >99.9%, are ideal for projects requiring high-confidence data. |
| Ultra-Long Reads |
Nanopore |
Capable of single-molecule reads up to megabase levels, effectively spanning complex genomic regions and reducing assembly difficulties. |
| Real-Time Data Analysis |
Nanopore |
Supports real-time data streaming, providing rapid results suitable for time-critical scenarios. |
| High Sample Throughput |
Nanopore |
The PromethION platform offers a throughput of 1.9 Tb per run, accommodating large-scale sample sequencing. |
4. Hybrid Sequencing Strategy
In certain scenarios, employing a hybrid sequencing strategy that integrates both PacBio and Nanopore technologies can maximize their respective advantages:
- Genome Assembly: Use Nanopore to generate ultra-long-read scaffolds, combined with PacBio HiFi reads for high-precision correction, enhancing assembly completeness and accuracy.
- Transcriptome Research: Leverage Nanopore for direct RNA sequencing to capture full-length transcripts, coupled with PacBio HiFi reads to validate alternative splicing events and RNA modifications.
Summary
When choosing between PacBio and Nanopore technologies, it is imperative to clarify research objectives (e.g., high precision, real-time capability), budgetary limits, and data requirements (e.g., read length, throughput). PacBio is particularly suitable for high-precision genome assembly and epigenetic studies, while Nanopore offers advantages in real-time monitoring, portability, and ultra-long-read applications. For complex projects, a hybrid sequencing strategy can offer a more comprehensive solution.
References:
- Hon, T., Mars, K., Young, G. et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data 7, 399 (2020). https://doi.org/10.1038/s41597-020-00743-4
- Udaondo, Zulema, et al. "Comparative analysis of PacBio and Oxford nanopore sequencing technologies for transcriptomic landscape identification of Penaeus monodon." Life 11.8 (2021): 862. https://doi.org/10.3390/life11080862
- Beckett, Angela H., Kate F. Cook, and Samuel C. Robson. "A pandemic in the age of next-generation sequencing." The Biochemist 43.6 (2021): 10-15. https://doi.org/10.1042/bio_2021_187
- Udaondo, Zulema, et al. "Comparative analysis of PacBio and Oxford nanopore sequencing technologies for transcriptomic landscape identification of Penaeus monodon." Life 11.8 (2021): 862. https://doi.org/10.3390/life11080862


 Sample Submission Guidelines
Sample Submission Guidelines
 Development of Long-read Sequencing Technology (2008-2012).
Development of Long-read Sequencing Technology (2008-2012). Flowchart of HiFi sequence read generation and downstream applications. (Hon, T., et al., 2020)
Flowchart of HiFi sequence read generation and downstream applications. (Hon, T., et al., 2020) A schematic diagram of the mechanism of Oxford Nanopore Technologies (ONT) sequencing. (Beckett, Angela H., et al., 2021)
A schematic diagram of the mechanism of Oxford Nanopore Technologies (ONT) sequencing. (Beckett, Angela H., et al., 2021) Experimental pipelines used in this study. Asterisk (*) indicates a step in the pipeline required for PacBio sequencing. (Udaondo, Zulema, et al. 2021)
Experimental pipelines used in this study. Asterisk (*) indicates a step in the pipeline required for PacBio sequencing. (Udaondo, Zulema, et al. 2021) Comparison of ONT and PacBio sequencing libraries. (Udaondo, Zulema, et al., 2021)
Comparison of ONT and PacBio sequencing libraries. (Udaondo, Zulema, et al., 2021)