The scientific domain of botanical genomics represents a dynamic and innovative research area investigating the complex genetic architecture of plant organisms. Cutting-edge molecular sequencing methodologies, including advanced next-generation and long-read technological platforms, have profoundly transformed scientific investigations into plant genetic structures. Researchers now possess unprecedented capabilities to analyze intricate genomic phenomena such as chromosome multiplication, transposable genetic elements, and genome expansion.
These technological breakthroughs have catalyzed significant agricultural advancements, enabling scientists to develop crop varieties with enhanced resilience to environmental stressors and improved nutritional profiles. CD Genomics play a crucial role in this scientific ecosystem, providing sophisticated sequencing and computational analysis services designed to unravel the intricate genetic potential inherent in plant biological systems.
What Are Plant Genomes?
Plant genomes represent the complete set of genetic material within a plant, including nuclear DNA and organelle genomes such as those in mitochondria and chloroplasts. They are responsible for encoding traits that determine a plant's growth, adaptability, and functionality.
Nuclear genome: This is the main part of the plant genome and is contained on the chromosomes within the nucleus. The nuclear genome is responsible for the transmission of most genetic information and is one of the key objects of research.
Mitochondrial genome: Mitochondria are important organelles responsible for energy metabolism in plant cells. Their genomes are relatively small and highly conserved, but have unique genetic mechanisms.
Chloroplast genome: Chloroplast is the place where plants conduct photosynthesis. Its genome is larger and more complex than the mitochondrial genome, and is usually inherited only from the female parent.
Service you may interested in
Resource
Diversity of Plant Genomes
The diversity of plant genomes is one of the most remarkable in nature, with an alarming range of sizes that can vary by more than 2400 times. For example, the genome size of Arabidopsis thaliana is about 135 Mb, while the genome size of Mexican white pine (Pinus ayacahuite) is more than 30 Gb.
Range and characteristics of plant genome size
Differences in plant genome size are mainly reflected in the accumulation of repetitive sequences, changes in the number of chromosomes, and the complexity of genome structure.
Smallest genome: Genlisea tuberosa is the smallest known plant genome, only 63 Mb in size, half the size of Arabidopsis.
Largest genome: Paris japonica is the largest plant genome currently sequenced, reaching 149 Gb, 50 times larger than the human genome.
Distribution of genome size: Plant genome size is extremely uneven, with most species having small genomes, while very large genomes occur only in certain groups.
This diversity influences plant functions such as photosynthesis, reproduction, and environmental adaptation. For example, chloroplast genomes, which encode genes essential for photosynthesis, are integral to understanding plant biology. Learn more about chloroplast DNA sequencing.
Why Are Plant Genomes So Large?
The reason why plant genomes are huge is mainly affected by polyploidy and whole-genome duplication (WGD). This phenomenon is common in many plant species, especially in crops such as wheat and corn.
Polyploidy and Whole-Genome Duplication
Polyploidy refers to cells containing more than two sets of chromosomes, while whole-genome replication refers to the replication of the entire genome. Both phenomena lead to significant increases in genome size. For example, crops such as wheat and corn gain additional copies of chromosomes through whole-genome replication, increasing the complexity and diversity of the genome.
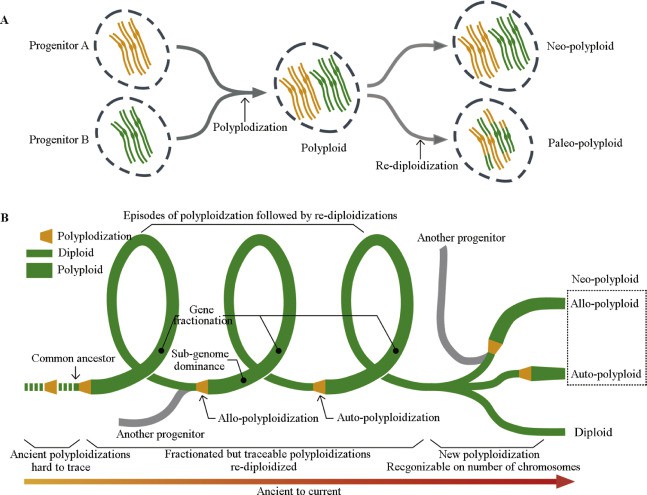 Figure 1. Life cycles of plants experiencing recurrent polyploidization and re-diploidization events(Kang ,et.al ,2019)
Figure 1. Life cycles of plants experiencing recurrent polyploidization and re-diploidization events(Kang ,et.al ,2019)
Genome-wide replication not only increases the number of genes, but also provides the basis for functional differentiation of genes. For example, in polyploid plants, many genes may be preserved for different biological functions, or may gradually lose function over the course of evolution. This expansion of the genome provides a greater genetic basis for plants to adapt to environmental changes, thereby improving their survival and reproduction capabilities.
Environmental and Evolutionary Factors
The expansion of plant genomes is also driven by environmental and evolutionary factors. In order to adapt to different climates and habitats, plants need to have greater genetic diversity and flexibility. For example, polyploid plants often have higher genetic variability and adaptability, which allows them to better cope with environmental pressures and challenges.
Transposon (TEs) are also one of the important factors affecting the size of plant genomes. These "jumping genes" are able to move through DNA and replicate themselves, significantly increasing the length of the genome. Studies have shown that transposons account for a significant proportion of many plant genomes, and their activity has a profound impact on genome structure and function.
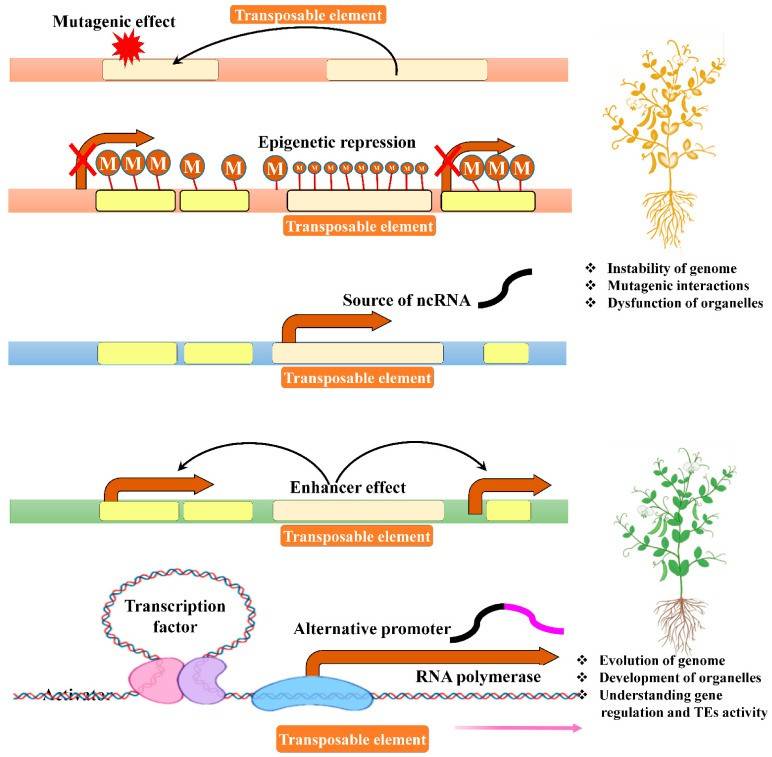 Figure 2.Primary regulatory roles of transposable elements(Ramakrishnan ,et.al ,2021)
Figure 2.Primary regulatory roles of transposable elements(Ramakrishnan ,et.al ,2021)
Although polyploidy and whole-genome duplication bring about genomic expansion, they also accompany a complex series of evolutionary processes. For example, many duplicate genes undergo sequence loss, silencing, or functional changes after replication. These changes not only affect the structure of the genome, but may also promote the creation of new functions or the improvement of existing functions.
Some studies have shown that polyploid plants may have experienced multiple genome-wide replication events during evolution and gradually simplified the genome through mechanisms such as genome fragmentation, gene loss, and non-functionalization. However, these simplified processes do not always completely eliminate redundant genes, but rather retain some potentially functional genes, thereby providing support for long-term adaptation of plants.
Comparative Genomics: Plants vs. Animals
There are significant differences in structure and evolution between plant genomes and animal genomes, and these differences reflect their different strategies in adapting to environmental changes and evolution.
Structural Differences
Genome size and complexity
Plant genomes are usually larger and more complex than animal genomes. For example, the genome size of wheat is more than five times that of humans. There are a large number of repetitive sequences, transposons (TEs), and genome-wide ploidy events in plant genomes. These factors lead to significantly higher dynamics and complexity in plant genomes than in animals. In contrast, animal genomes are small and relatively stable, with relatively conservative chromosome numbers and genomic structure.
Chromosomes and repetitive sequences
Genome-wide polyploidy events occur frequently in plant genomes, which gives plant chromosome number and genome size a greater range of variation. For example, certain plants may have three or more sets of chromosomes. In addition, transposons are active in plant genomes and can change the structure of the genome through insertions, deletions, and recombination. In contrast, animal genomes have fewer repetitive sequences and lower frequency of ploidy events.
Mitochondrial and chloroplast genomes
Plant genomes also include unique chloroplast and mitochondrial genomes, which play important roles in plant evolution. For example, plant mitochondrial genomes are much larger in size than animal mitochondrial genomes, and their structure and function are highly diverse. In contrast, animal mitochondrial genomes are small and relatively conservative.
Evolutionary Insights
Plasticity and adaptability of the genome
The high plasticity of plant genomes allows them to quickly adapt to environmental changes. For example, plants respond to environmental stress through genomic ploidy events, transposon regulation, and epigenetic mechanisms. This flexibility allows plants to show greater adaptability and resilience in the face of challenges such as climate change.
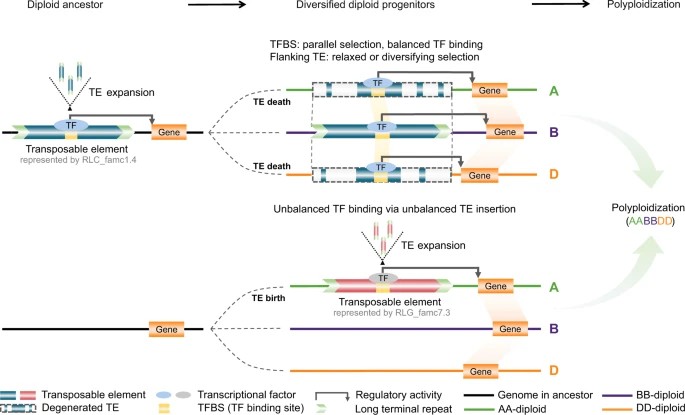 Figure 3.The effects of TE-derived regulatory conservation and innovation.(Zhang ,et.al ,2022)
Figure 3.The effects of TE-derived regulatory conservation and innovation.(Zhang ,et.al ,2022)
Evolutionary dynamics and diversity
The dynamics of plant genomes are not only reflected in structure, but also in function. For example, silencers and other regulatory elements in plant genomes are reused during the insertion of repetitive sequences, thereby promoting the creation of new functions. In contrast, the evolution of animal genomes relies more on gradual changes and maintenance of conservation.
Environmental adaptation and survival strategies
Plants respond to the environmental challenges posed by immobility through their dynamic genomic structure and function. For example, plants adapt to environmental pressures such as drought and cold through complex signaling and metabolic pathways. In contrast, animals respond to environmental changes through the coordinated role of the nervous and immune systems.
Arabidopsis thaliana: A Model Plant for Genome Analysis
Arabidopsis thaliana is a widely studied model plant. Because of its small genome, short life cycle, and easy genetic manipulation, it has become an important model in plant genomics research. The following is a detailed analysis of the importance and contribution of Arabidopsis in genomic research:
Importance in Research
The genome is small and simple in structure: The Arabidopsis genome is approximately 125-157 Mb in size and contains five chromosomes, making it an ideal target for studying the structure and function of the genome. Its relatively small genome simplifies genetic analysis and gene mapping.
Rapid life cycle: It takes only about six weeks from seed germination to plant maturity, allowing scientists to complete multiple generations of experiments in a short period of time, accelerating genetic research.
Self-pollinated and easy to cultivate: Arabidopsis is a self-pollinated plant with high seed yields and easy to preserve, making it suitable for large-scale laboratory cultivation and genetic experiments.
Abundant mutant resources: Arabidopsis has a large number of known mutants, which provide valuable tools for studying gene function.
Contributions to Plant Genomics
Gene regulation mechanism:
The Arabidopsis genome sequencing project was completed in 2000 and revealed the functions of approximately 25,000 genes, laying the foundation for understanding plant gene regulation mechanisms. Genomic information facilitates the study of gene expression regulatory networks, such as the interpretation of transcription factors and epigenetic regulatory mechanisms.
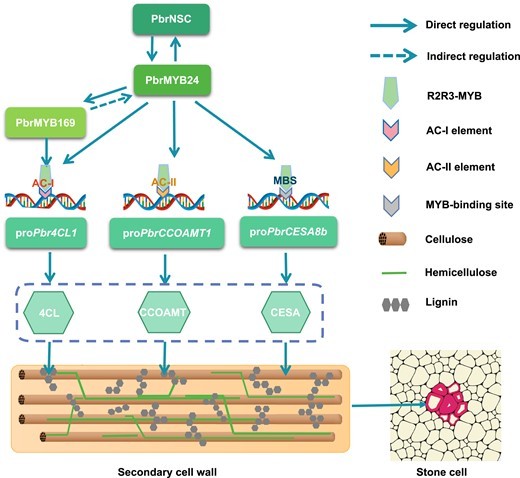 Figure 4.The hypothetical working model of PbrMYB24 regulation of stone cell formation.(Xue, Y,et.al ,2023)
Figure 4.The hypothetical working model of PbrMYB24 regulation of stone cell formation.(Xue, Y,et.al ,2023)
Plant development pathways:
Arabidopsis is used to study key processes in plant growth and development, including seed formation, root development and flower organ development. Its model status promotes the understanding of developmental processes such as plant hormone signaling, cell division and differentiation.
Environmental response:
Arabidopsis is widely used to study plant responses to environmental stresses, including drought, salt stress and pathogen attack. Its rapid growth and genetic manipulation capabilities make it an ideal model for studying plant adaptive evolution and stress resistance.
Because of its unique biological characteristics, Arabidopsis plays an irreplaceable role in plant genomics research. Its small and compact genome, rapid life cycle and abundant mutant resources make it an ideal model for studying plant genetics, developmental biology and environmental responses. In addition, genomic data of Arabidopsis not only promotes basic scientific research, but also provides important technical support for agricultural production and crop improvement.
Current Trends in Plant Genomics
Plant genomics has made remarkable progress in recent years, mainly in the fields of technological innovation, agricultural applications, and biodiversity protection.
Technological Advancements
Next-generation sequencing technology (NGS) and third-generation sequencing technologies such as PacBio SMRT and Oxford Nanopore have greatly promoted the development of plant genomics. These technologies can generate longer reads and improve the accuracy and speed of genome assembly while reducing costs.
High-throughput sequencing platforms such as Illumina play an important role in plant genome research, making large-scale genome sequencing possible. These platforms not only reduce sequencing costs, but also improve data production efficiency and provide strong technical support for plant genomics research.
With the advancement of sequencing technology, bioinformatics tools are also constantly developing. For example, tools used to process NGS data (such as Poretools, Genopo, etc.) greatly improve the efficiency and accuracy of data analysis. In addition, NGS-based data analysis methods such as genome-wide association studies and QTL mapping have further deepened the understanding of plant genetic characteristics.
Applications in Agriculture
Development of pest-resistant crops
Plant genomics provides a scientific basis for cultivating pest-resistant crops by accurately locating genes related to insect resistance. For example, using SNP markers and QTL analysis, researchers can quickly identify insect resistance traits and apply them in breeding
Improve crop stress resistance
Genomic technology helps scientists study the response mechanisms of crops to environmental stresses such as drought and high temperatures. By integrating genomic selection (GS) and phenotypic prediction, researchers were able to develop crop varieties that were more tolerant to stress.
Improve yield and nutritional value
Breeding strategies based on genome selection have been shown to significantly improve crop yield and nutritional value. For example, by analyzing genetic regions associated with yield, researchers can design high-yield and nutrient-rich crops.
Biodiversity Conservation
Plant genomics technology plays an important role in the protection of endangered species. By sequencing the genomes of endangered plants, researchers are able to identify their unique genetic characteristics and take effective measures to protect them.
Conclusion
Plant genomics is at the forefront of addressing global challenges like food security, environmental conservation, and sustainable agriculture. As a trusted partner in genomics research, CD Genomics offers cutting-edge sequencing and bioinformatics solutions tailored to plant genomics studies.
References:
- Kang Zhang, Xiaowu Wang, Feng Cheng, Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication,Horticultural Plant Journal,Volume 5, Issue 6,2019,Pages 231-239, https://doi.org/10.1016/j.hpj.2019.11.003.
- Ramakrishnan, M., Satish, L., Kalendar, R., et.al. (2021). The Dynamism of Transposon Methylation for Plant Development and Stress Adaptation. International journal of molecular sciences, 22(21), 11387. https://doi.org/10.3390/ijms222111387
- Zhang, Y., Li, Z., Liu, J. et al. Transposable elements orchestrate subgenome-convergent and -divergent transcription in common wheat. Nat Commun 13, 6940 (2022). https://doi.org/10.1038/s41467-022-34290-w
- Xue, Y., Shan, Y., Yao, J. L., et.al. (2023). The transcription factor PbrMYB24 regulates lignin and cellulose biosynthesis in stone cells of pear fruits. Plant physiology, 192(3), 1997–2014. https://doi.org/10.1093/plphys/kiad200


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Life cycles of plants experiencing recurrent polyploidization and re-diploidization events(Kang ,et.al ,2019)
Figure 1. Life cycles of plants experiencing recurrent polyploidization and re-diploidization events(Kang ,et.al ,2019) Figure 2.Primary regulatory roles of transposable elements(Ramakrishnan ,et.al ,2021)
Figure 2.Primary regulatory roles of transposable elements(Ramakrishnan ,et.al ,2021) Figure 3.The effects of TE-derived regulatory conservation and innovation.(Zhang ,et.al ,2022)
Figure 3.The effects of TE-derived regulatory conservation and innovation.(Zhang ,et.al ,2022) Figure 4.The hypothetical working model of PbrMYB24 regulation of stone cell formation.(Xue, Y,et.al ,2023)
Figure 4.The hypothetical working model of PbrMYB24 regulation of stone cell formation.(Xue, Y,et.al ,2023)