What is Chloroplast DNA (cpDNA) Sequencing
Chloroplasts are organelles found in plant cells and eukaryotic algae that conduct photosynthesis,they are not only essential for the life of plants but for all life on Earth. Chloroplasts, like mitochondria, contain their own DNA, comprising approximately 130 genes, which are involved in photosynthesis and other important metabolic processes. Chloroplast genomes display considerable variation both within and between species, providing insights into the phylogeny and evolutionary adaption. Chloroplasts also carry out a number of other functions, including fatty acid synthesis, much amino acid synthesis, and the immune response in plants.
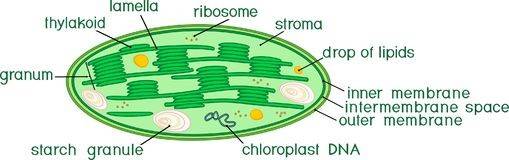
The chloroplast DNA (cpDNA) sequencing is a high-throughput sequencing of plant chloroplast genomes using Illumina and PacBio platforms to perform an in-depth analysis of sequence information. Through comparative genomic analysis, information such as species classification, phylogenetic evolution, geographic lineage inheritance, disease diagnosis and forensic science is obtained, revealing its important role in species origin, biological evolution and genetic relationship between different species.
If you want to learn more about chloroplasts, you can refer to our article "Chloroplast Fact Sheet: Definition, Structure, Genome, and Function."
Advantages of Our cpDNA Sequencing Service
- Flexible experimental protocol: adopt appropriate sequencing solutions according to different needs
- Strong bioinformatics team: in addition to providing standard bioinformatics services, it also provides personalized bioinformatics services
- Experienced: chloroplast sequencing of multiple species has been completed
- Cost-effective and time-efficient
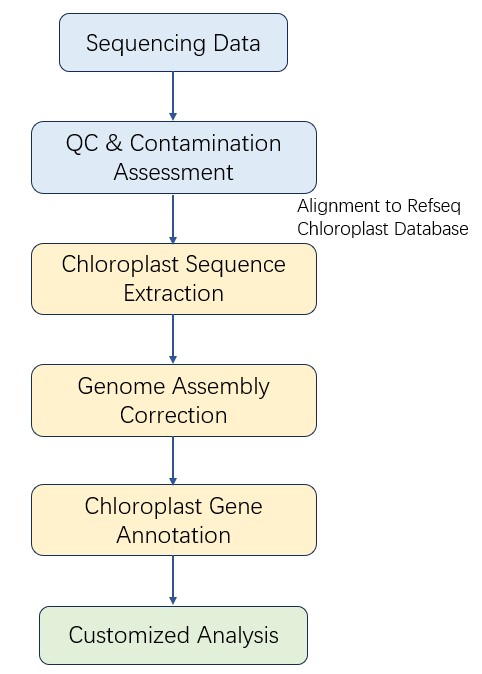
Chloroplast DNA (cpDNA) Sequencing Workflow
CD Genomics employs multiple platforms to provide the fast and accurate Chloroplast DNA (cpDNA) Sequencing services and bioinformatics analysis. Our highly experienced experts execute quality management, following every procedure to ensure high quality results. The general workflow for Chloroplast DNA (cpDNA) Sequencing is outlined below.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategies
|
Data Analysis
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Chloroplast DNA (cpDNA) Sequencing for your writing (customization)
CD Genomics offers cost-effective protocol of cpDNA sequencing, including DNA preparation, chloroplast enrichment, sequencing, and analyze the entire chloroplast genome from total DNA samples. We can tailor this pipeline to your research interest. If you have additional requirements or questions, please feel free to contact us.
1. Why Conduct Chloroplast DNA Sequencing?
Chloroplast DNA sequencing plays a pivotal role in various realms such as plant taxonomy, phylogenetic analyses, population genetics, biodiversity conservation, and agronomic improvement. This methodology empowers us to illuminate the intricate evolutionary connections within plant species and explore genetic diversity, facilitating the identification and enhancement of superior plant varieties.
2. How Does Chloroplast DNA Differ from Nuclear DNA?
Chloroplast DNA, a compact circular molecule ensconced within the chloroplast, chiefly orchestrates vital processes like photosynthesis. In contrast, nuclear DNA abides within the cell nucleus, serving as the primary genetic reservoir in plants housing the majority of genes and genetic data.
3. How to Extract Chloroplast DNA?
The extraction of chloroplast DNA entails the isolation of chloroplasts from plant tissues, succeeded by the extraction of DNA from these organelles. Techniques commonly employed include mechanical disruption of plant leaves, differential centrifugation for chloroplast isolation, and DNA purification through methods such as CTAB or dedicated commercial kits.
4. What are the Common Chloroplast DNA Sequencing Methods?
Common sequencing methods include traditional Sanger sequencing and next-generation sequencing (NGS) technologies like Illumina, PacBio, and Oxford Nanopore. NGS technologies have emerged as the mainstream choice for chloroplast DNA sequencing due to their high throughput and relatively lower costs.
5. How to conduct quality control and contamination assessment?
Quality control steps include using software such as FastQC to check the read length and quality of sequencing data, and removing low-quality reads. Contamination assessment involves aligning reads to known databases to detect possible exogenous DNA contamination.
6. What is personalized analysis?
Personalized analysis is a customized analysis of sequencing data based on specific research needs. For example, in-depth study can be conducted on specific genes, or differences in chloroplast genomes of different samples can be compared.
7. What is Refseq chloroplast database alignment?
Refseq chloroplast database alignment is aligning sequencing data to known chloroplast genome sequences in the Refseq database for gene annotation, variant detection, and phylogenetic analysis. This helps to confirm the accuracy and completeness of sequencing results.
8. What are the applications of chloroplast DNA sequencing?
- Phylogenetics: Unraveling the evolutionary relationships among plants.
- Population Genetics: Investigating genetic diversity within populations and gene flow.
- Conservation Biology: Formulating conservation strategies to safeguard endangered species.
- Agriculture and Breeding: Selecting and improving superior crop varieties for agricultural advancement.
These applications underscore the versatility and significance of chloroplast DNA sequencing in elucidating evolutionary, ecological, and agricultural phenomena.
Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops
Journal: Scientific Reports
Impact factor: 4.6
Published: 14 December 2022
Background
The genus Vaccinium includes over 450 species with major commercial crops such as blueberries, cranberries, bilberries, and lingonberries. These berries are economically important and widely consumed for their nutritional value. Taxonomic classification within Vaccinium is complex due to phenotypic variability and hybridization. Chloroplast genome sequencing offers potential for better taxonomic resolution, breeding, biotechnology, and product authentication. This study reports new chloroplast genomes for key Vaccinium species, providing valuable resources for future research and practical applications.
Methods
- V. corymbosum hybrid (SHB)
- V. virgatum (RB)
- V. angustifolium (LB)
- DNA extraction
- DNA library preparation
- PacBio sequencing
- Illumina whole genome sequencing
- Assembly and polishing
- Gene annotation
- Comparative analyses
- SSRs and indel detection
Results
Chloroplast genomes for five Vaccinium species were assembled using PacBio long reads for SHB and rabbiteye, resulting in complete gapless sequences. For NHB, lowbush, and bilberry, Illumina short reads were used, yielding assemblies with some gaps. Final cpDNA sequences exhibited a quadripartite structure, including large and small single copy regions, and a pair of inverted repeats.
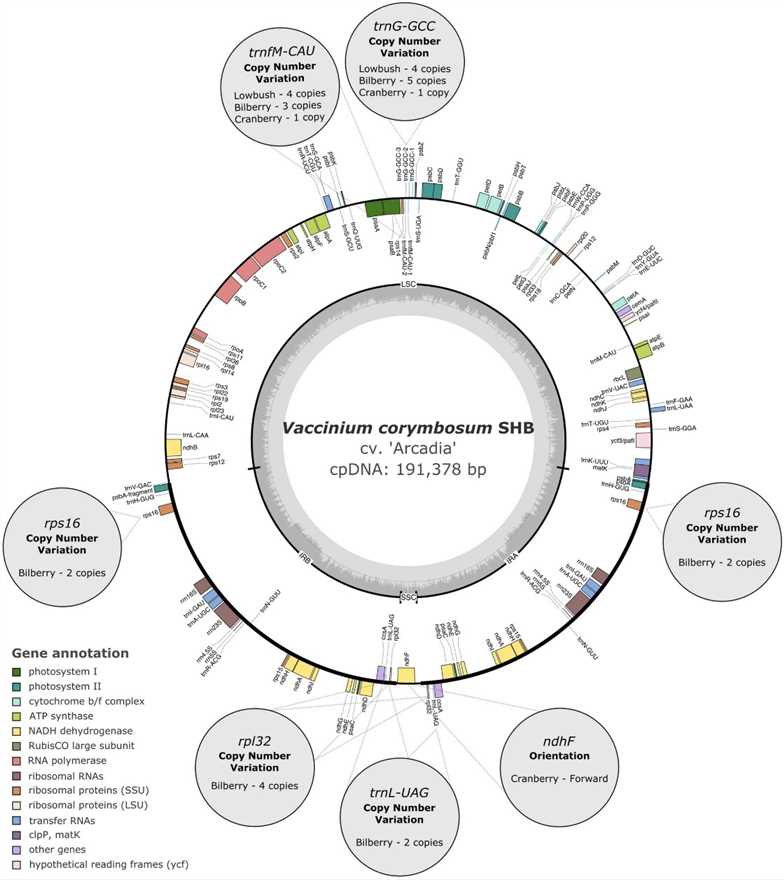 Fig 1. Circular chloroplast genome map of southern highbush blueberry cv. 'Arcadia' (V. corymbosum hybrids).
Fig 1. Circular chloroplast genome map of southern highbush blueberry cv. 'Arcadia' (V. corymbosum hybrids).
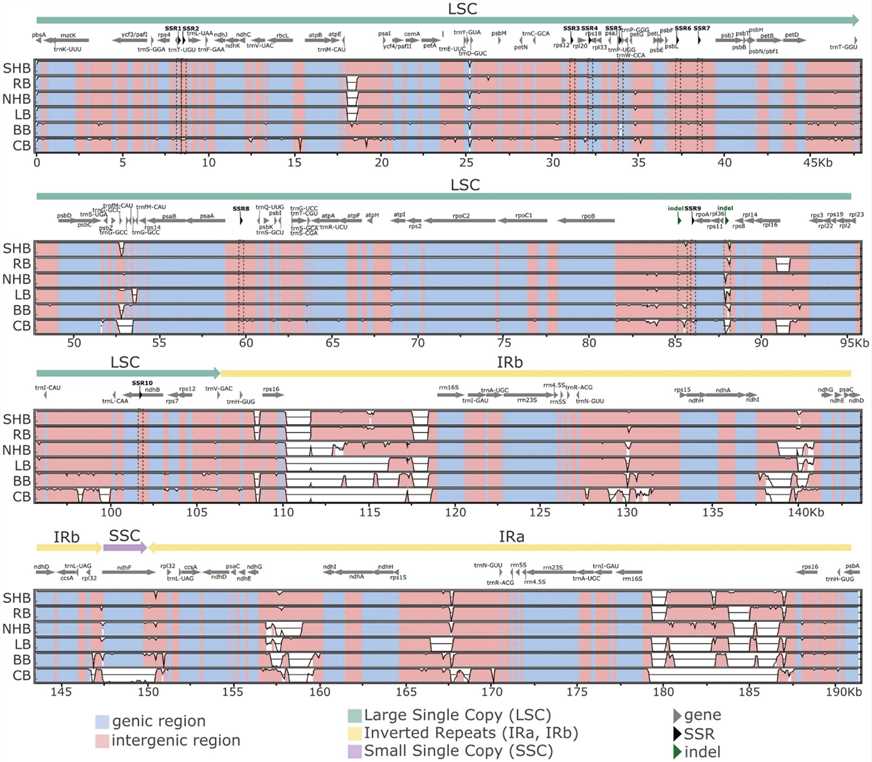 Fig 2. Multiple sequence alignment of Vaccinium chloroplast genomes performed with mVISTA.
Fig 2. Multiple sequence alignment of Vaccinium chloroplast genomes performed with mVISTA.
The chloroplast DNA (cpDNA) of five Vaccinium species (SHB, NHB, rabbiteye, lowbush, and bilberry) was annotated alongside the cranberry cpDNA from GenBank. Each cpDNA contained 112 unique functional genes, with some variability in gene copy numbers. Most genes were found in the large single copy (LSC) region, while the small single copy (SSC) and inverted repeats (IRs) contained fewer genes.
Comparative analysis showed high conservation and synteny among the cpDNAs, with most differences in non-coding regions and around IR borders. Simple sequence repeats (SSRs) and insertion-deletion polymorphisms were analyzed, identifying 10 polymorphic SSR loci useful for species differentiation. Two specific markers, "rpl36-rps8" and "rpoB-rpoA (3)," were identified to discriminate all six commercially relevant Vaccinium species.
Conclusion
This study assembled chloroplast genomes of five key Vaccinium species, revealing high conservation but some variability in non-coding regions. Comparative analyses identified regions useful for distinguishing these species and highlighted evolutionary patterns within highbush blueberries. More chloroplast genomes will aid further study and understanding of Vaccinium crop evolution.
Reference:
- Fahrenkrog, A.M., Matsumoto, G.O., Toth, K. et al. Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops. Sci Rep 12, 21600 (2022).
Here are some publications that have been successfully published using our services or other related services:
Rubisco from two ecotypes of Plantago lanceolata L. that are native to sites differing in atmospheric CO2 levels
Journal: bioRxiv
Year: 2024
High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing
Journal: International Journal of Molecular Sciences
Year: 2020
Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI
Journal: New phytologist
Year: 2017
Generation of a highly attenuated strain of Pseudomonas aeruginosa for commercial production of alginate
Journal: Microbial Biotechnology
Year: 2019
Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Journal: Viruses
Year: 2024
Genome Analysis and Replication Studies of the African Green Monkey Simian Foamy Virus Serotype 3 Strain FV2014
Journal: Viruses
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
