What is Low-Pass Whole Genome Sequencing
Low-pass whole-genome sequencing (LP-WGS) is a method involving high-throughput sequencing of entire genomes at relatively low sequencing depths. Typically, LP-WGS operates at a depth of 0.5-1× coverage, meaning each base pair is sequenced, on average, between 0.5 to 1 time. The approach compensates for these sparse data points by employing computational techniques, such as genotype imputation algorithms, to extrapolate missing sequences, thereby delivering a relatively comprehensive genomic landscape.
As an economical and efficient genomic sequencing strategy, LP-WGS is particularly suited for extensive population studies and the calculation of polygenic risk scores. By lowering sequencing depth and utilizing computational genotype imputation, the method significantly reduces costs and data storage requirements, while still delivering genome information that, in certain contexts, stands on par with traditional whole-genome sequencing (WGS).
Technical Foundations and Distinctions from Traditional WGS
1. Sequencing Depth:
- Traditional WGS typically necessitates high sequencing depths (e.g., 30× or more) to ensure multiple readings of each base, thus achieving high precision in genomic data.
- In contrast, LP-WGS utilizes reduced sequencing depths (0.5×-1×), inferring genotypes and filling in gaps through computational methodologies.
2. Cost and Data Volume:
- LP-WGS markedly reduces sequencing costs and the need for data storage, as its lower sequencing depth demands less time and significantly diminishes storage space requirements.
- Traditional WGS, while offering more comprehensive genomic insights, entails higher costs and greater data processing and storage overheads.
3. Applications:
- LP-WGS finds its applications in large-scale population studies, genome-wide association studies (GWAS), and polygenic risk score computations.
- Traditional WGS is extensively employed in genetic disorder diagnostics, cancer research, and microbiome analysis.
4. Data Quality:
- LP-WGS, relying on computational genotype inference, may introduce certain errors and uncertainties. Therefore, enhancing accuracy requires integrating probabilistic frameworks and leveraging large-scale sample data.
- Given its high sequencing depth, traditional WGS offers superior data quality, making it suitable for studies demanding high-precision genomic information.
Advantages and Applications of LP-WGS
Low-Pass Whole Genome Sequencing stands out as a cost-effective and efficient genome sequencing technology, offering significant promise across various applications. Herein, we explore the advantages and potential applications of LP-WGS:
Advantages of Low Sequencing Depth
1. Cost-Effectiveness:
- A primary advantage of LP-WGS is its cost efficiency. By utilizing low sequencing depths, LP-WGS significantly reduces costs, positioning itself as a valuable tool in genetic diagnostics, especially beneficial in low- and middle-income countries.
- Compared with traditional chromosomal microarray analysis (CMA), LP-WGS incurs only a quarter of the cost, yet provides reliable copy number variation (CNV) detection. This affordability is particularly crucial in low-income settings, expanding access to genetic testing for numerous patients. Moreover, LP-WGS offers a higher degree of automation, minimizing technical errors and reducing the demand for DNA input.
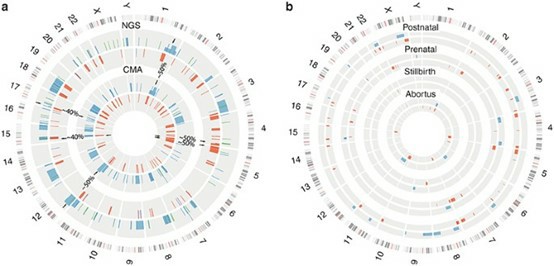 CNV detection in a validation group and a multicenter clinical group. (Dong, Z., et al., 2016)
CNV detection in a validation group and a multicenter clinical group. (Dong, Z., et al., 2016)
2. Data Processing and Storage:
- Due to its reduced data volume, LP-WGS decreases the requirements for data processing and storage, thereby conserving computational resources.
- Techniques such as genotype imputation effectively fill gaps in sequencing data, presenting a more comprehensive genomic overview.
3. Flexibility and Scalability:
- LP-WGS can decrease costs further by increasing the number of samples processed while maintaining high data quality.
- Although traditional WGS delivers higher data quality, its elevated costs restrict its application in large-scale population studies.
4. High Throughput:
- Capable of processing large sample volumes, LP-WGS lends itself well to extensive research undertakings.
5. Comprehensiveness:
- Despite the lower coverage, LP-WGS successfully identifies a substantial array of CNVs and single nucleotide variants (SNVs).
Applications in Clinical Cytogenetics
LP-WGS demonstrates robust performance in clinical cytogenetics, particularly for identifying chromosomal abnormalities and CNVs. Studies indicate that LP-WGS exhibits sensitivity and accuracy comparable to CMA in detecting fetal chromosomal abnormalities, cases of high-risk pregnancies, infertility, and congenital malformations. For example, research conducted in Brazil underscored the comparable efficacy of LP-WGS to CMA in identifying fetal chromosomal anomalies, with added cost benefits.
Potential in Large-Scale Population Genomics
LP-WGS showcases immense potential in expansive population genomic studies. Its lower cost and higher throughput facilitate genome-wide association studies (GWAS) and whole-genome selection studies, aiming to uncover disease-associated genes and variations. Leveraging genotype imputation techniques, LP-WGS provides high-density genetic markers, suitable for the analysis of large sample cohorts.
Clinical and Research Case Studies
Multiple studies and practical cases underscore the significant efficacy of LP-WGS in both clinical and research settings. For instance, in prostate cancer (PCa) patients, LP-WGS has been employed to detect circulating tumor DNA (ctDNA) percentages and CNVs, offering crucial insights into disease prognosis. Additionally, LP-WGS has proven advantageous in identifying CNVs in early-stage PCa patients, highlighting its cost-effectiveness.
Broad Spectrum of Applications
LP-WGS finds extensive applications, including but not limited to:
- Cancer Diagnostics: Detecting CNVs and ctDNA for early diagnosis and prognosis evaluation in cancer.
- Genetic Disease Diagnosis: Identifying chromosomal abnormalities and CNVs for the diagnosis of genetic disorders and prenatal testing.
- Population Genomics: Facilitating large-scale population studies to unveil genetic diversity and its association with diseases.
- Agriculture and Breeding: Applied in trait association and breeding evaluations for species with large genomes.
Services you may interested in
Want to know more about the details of LP-WGS? Check out these articles:
The Human LP-WGS Market: Current Status and Trends
The market for Low-Pass Whole Genome Sequencing is undergoing rapid expansion, marked by its potential applications across various domains such as neonatal screening, cancer detection, genetic disease diagnosis, and routine health assessments. Technological advancements coupled with cost reductions position LP-WGS as a pivotal tool for precision medicine and personalized treatments in the coming years.
1. Market Dynamics of LP-WGS
The LP-WGS market has experienced significant growth, largely attributed to its cost-effectiveness and high-throughput capabilities. Conventional Whole Genome Sequencing (WGS), with costs running into thousands of dollars for 30x coverage, contrasts sharply with LP-WGS, which lowers sequencing depth (typically 0.1x-1x) to reduce costs below $200 per sample while retaining high sensitivity in detecting copy number variations (CNVs). For instance, automated library preparation technologies developed by companies like MGI enable the processing of 1,536 samples in a single day, with an annual sequencing throughput reaching up to 384,000 samples at 1.0x coverage. This cost advantage has propelled its rapid adoption in clinical diagnostics and large-scale population studies.
2. Current Applications in Genomics and Clinical Diagnostics
LP-WGS has found extensive applications in several key areas:
- Prenatal Diagnosis: Utilizing low coverage sequencing of 15M reads per sample, LP-WGS effectively detects chromosomal aneuploidies (such as trisomy 21) and microdeletion syndromes with a sensitivity of 99%, and a sample repeat experiment rate of just 0.5% (much lower than CMA's 4.6%).
- Cancer Genomics: When integrated with deep learning models like GIInger, LP-WGS can identify genomic instability resulting from homologous recombination defects (HRD) and predict the sensitivity of ovarian and breast cancer patients to PARP inhibitors, achieving a 98% consistency with high-depth sequencing results.
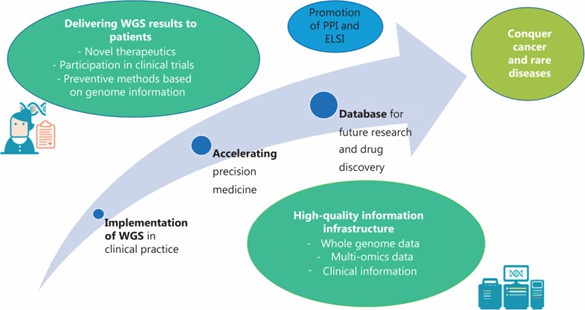 An Overview of Japan's "Action Plan for Whole Genome Analysis of Cancer and Rare/Intractable Diseases" WGS Initiative (Yuki Katsuya, 2024)
An Overview of Japan's "Action Plan for Whole Genome Analysis of Cancer and Rare/Intractable Diseases" WGS Initiative (Yuki Katsuya, 2024)
- Population Genetics: In studies of isolated populations like the Pacific Kosraeans, LP-WGS enables the determination of genetic variations across 60% of the population through genotype imputation from just seven samples at 5x coverage, with an accuracy exceeding 97%.
3. Emerging Market Trends and Commercial Adoption
The commercial adoption of LP-WGS is characterized by several trends:
- Technological Integration: Emerging techniques such as Blended Genome-Exome Sequencing (BGE), which combines low-depth WGS (1-4x) with exome sequencing (30-40x), facilitate the concurrent detection of common genomic variants and rare exome-region mutations. This approach is a focal development area for 2024.
- Automation Enhancements: Clinical laboratories are refining standardized processes, such as PCR-free library preparation, to reduce the experimental cycle to seven days, with an initial DNA input of just 50ng, thereby significantly enhancing detection efficiency.
- Industry Collaboration: Genetic technology companies are advancing the integration of LP-WGS with artificial intelligence analysis platforms to optimize data interpretation capabilities and expand clinical applications.
4. Future Potential of LP-WGS
LP-WGS holds considerable promise in several areas:
- Neonatal Screening: Its low cost and high throughput make it ideal for large-scale screening of congenital genetic disorders like spinal muscular atrophy, with the potential to replace traditional biochemical testing methods.
- Cancer Early Detection: By identifying low-frequency CNVs in circulating tumor DNA (ctDNA), LP-WGS offers prospects for early cancer diagnosis and relapse monitoring.
- Genetic Disease Diagnosis: In populations with developmental delays or rare diseases, LP-WGS can screen hundreds of pathogenic CNVs simultaneously, addressing gaps left by SNP chips in genomic "blind spots."
Future Outlook for LP-WGS
The future trajectory of Low-Pass Whole Genome Sequencing reflects its potential to significantly advance personalized medicine and genetic disease research through multifaceted avenues.
Technological Advancements and Cost Reduction
With the relentless progression of sequencing technologies, particularly the advancements in Next-Generation Sequencing (NGS) platforms, the expense associated with LP-WGS is plummeting. This cost reduction enhances the feasibility of large-scale genomic projects and the implementation of personalized medicine approaches. For instance, the introduction of sequencing instruments such as the T20 by BGI offers substantial cost efficiency for whole genome sequencing, rendering it more accessible. Furthermore, trends in technology miniaturization, parallel sequencing techniques, and improvements in matrix reading methods are expected to facilitate further decreases in sequencing costs.
Applications in Personalized Medicine
The potential applications of LP-WGS in personalized medicine are vast. Whole genome sequencing enables physicians to garner comprehensive insights into an individual's genetic makeup, facilitating tailored therapeutic strategies. For example, by analyzing a patient's genomic data, healthcare professionals can predict drug responses, allowing for the optimization of medication dosages and treatment protocols. Moreover, LP-WGS can identify individuals at high risk for certain conditions, thereby supporting strategies for early intervention and prevention.
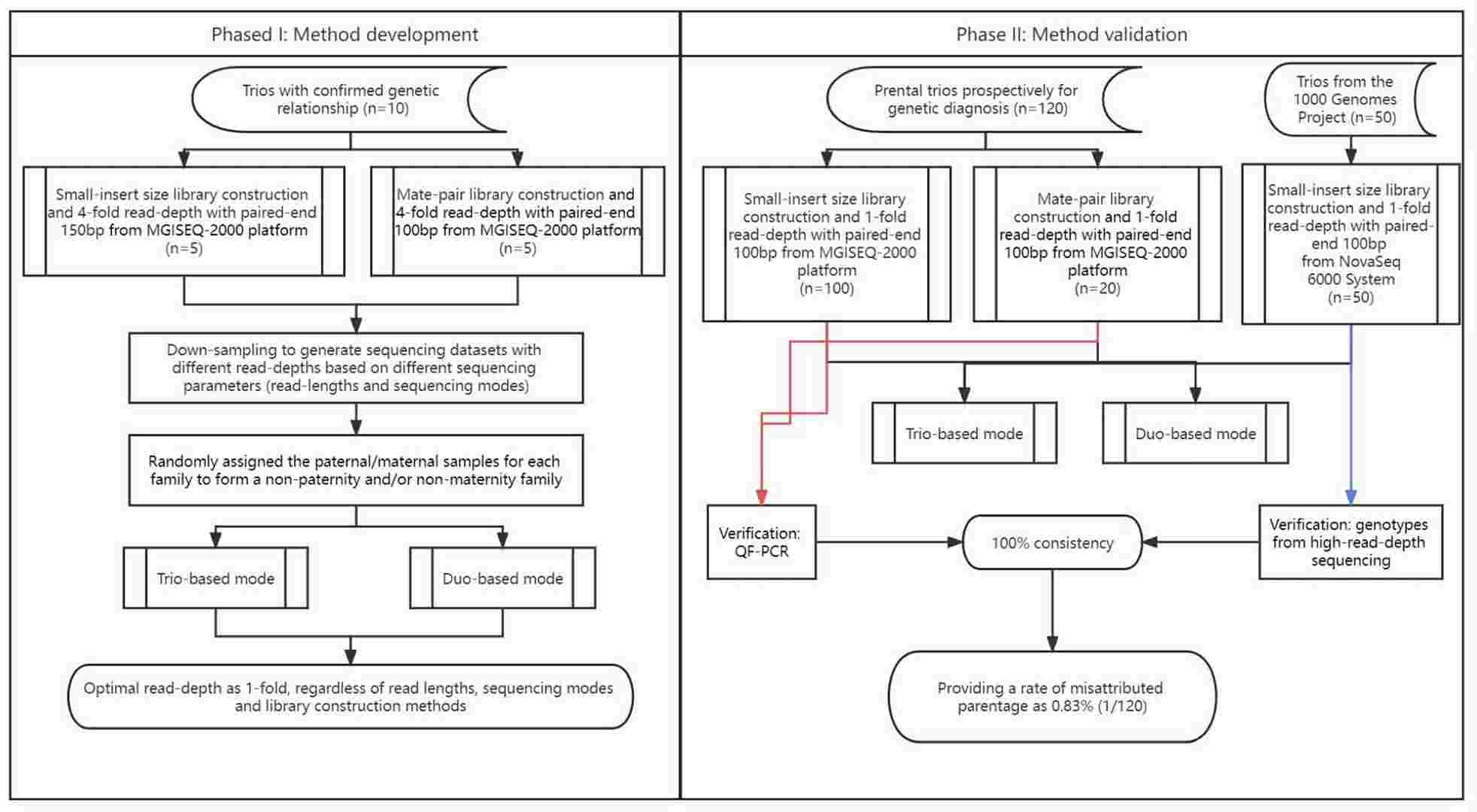 Workflow of method development and evaluation with low-pass genome sequencing for the paternity test. (Li, Keying, et al., 2023)
Workflow of method development and evaluation with low-pass genome sequencing for the paternity test. (Li, Keying, et al., 2023)
Role in Genetic Disease Research
In the realm of genetic disease research, LP-WGS holds tremendous value. Through comprehensive genome sequencing, researchers can identify disease-associated genetic variants more effectively, expediting the discovery of pathogenic genes and therapeutic targets. For instance, in diagnosing rare genetic disorders, whole genome sequencing offers superior diagnostic utility over chromosomal microarray analysis. Additionally, LP-WGS facilitates large-scale population genomic studies, contributing to a deeper understanding of genetic diversity across different populations.
Insights into Complex Genetic Disorders
LP-WGS offers a unique advantage in elucidating complex genetic disorders. Such conditions often involve intricate interactions among multiple genes, which traditional genetic testing methods may not thoroughly address. Whole genome sequencing provides a holistic view of the genetic landscape, allowing researchers to analyze genome characteristics comprehensively and enhance understanding of disease pathogenesis. For example, certain cancers may involve mutations across various genes implicated in cellular regulation, for which LP-WGS can offer a complete perspective.
Future Prospects
As sequencing costs continue to decline and technological innovations advance, LP-WGS is poised for broader application in both clinical and research settings. This will catalyze the development of personalized medical approaches and deepen investigations into genetic diseases. Moreover, the adoption of LP-WGS is anticipated to enhance public health by facilitating early diagnostics and personalized interventions, thereby improving patient quality of life.
Conclusion
Low-Pass Whole Genome Sequencing leverages low sequencing depth (0.1x-1x) in conjunction with genotype imputation techniques to detect large-scale chromosomal variations, such as copy number variations (CNVs) and microdeletions, with high sensitivity at just one-thirtieth the cost of traditional sequencing methods. This efficient approach has positioned LP-WGS as a transformative tool in both clinical and research domains. Its advantages include reduced cost (less than $200 per sample), high throughput (processing over 380,000 samples annually), and extraordinary flexibility, making it suitable for prenatal diagnosis, early cancer screening, and population genetics research.
Clinically, LP-WGS excels in the detection of prenatal chromosomal abnormalities with a sensitivity greater than 99%, as well as in the genomic analysis of cancers-such as the identification of homologous recombination deficiencies (HRD)-and in the screening of genetic disorders. The automation of LP-WGS processes further lowers the technological barriers to its adoption, facilitating broader accessibility and implementation.
Looking ahead, advancements in algorithmic optimization (e.g., haplotype reference panels) and technology integration (e.g., blended genome-exome sequencing) are poised to enhance the impact of LP-WGS even further. These innovations will promote the widespread applicability of precision medicine, establishing LP-WGS as a core driver in areas such as neonatal screening, the elucidation of complex diseases, and extensive public health initiatives. Consequently, LP-WGS is anticipated to expedite the comprehensive integration of genomic medicine from the research sphere into clinical practice.
References:
- Li, Jeremiah H., et al. "Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays." Genome research 31.4 (2021): 529-537. https://www.genome.org/cgi/doi/10.1101/gr.266486.120.
- DeSaix, Matthew G., et al. "Low‐coverage whole genome sequencing for highly accurate population assignment: Mapping migratory connectivity in the American Redstart (Setophaga ruticilla)." Molecular Ecology 32.20 (2023): 5528-5540. https://doi.org/10.1111/mec.17137
- Mazzonetto, Patricia C., et al. "Low‐pass whole genome sequencing as a cost‐effective alternative to chromosomal microarray analysis for low‐and middle‐income countries." American Journal of Medical Genetics Part A 194.11 (2024): e63802. https://doi.org/10.1002/ajmg.a.63802
- Wang, Ting, et al. "Expanded newborn screening for inborn errors of metabolism by tandem mass spectrometry in Suzhou, China: disease spectrum, prevalence, genetic characteristics in a Chinese population." Frontiers in genetics 10 (2019): 1052. https://doi.org/10.3389/fgene.2019.01052
- Katsuya, Yuki. "Current and future trends in whole genome sequencing in cancer." Cancer Biology & Medicine 21.1 (2024): 16. doi: 10.20892/j.issn.2095-3941.2023.0420
- Dong, Z., Zhang, J., Hu, P. et al. Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med 18, 940–948 (2016). https://doi.org/10.1038/gim.2015.199
- Li, Keying, et al. "Low-pass genome sequencing-based detection of paternity: validation in clinical cytogenetics." Genes 14.7 (2023): 1357. https://doi.org/10.3390/genes14071357


 Sample Submission Guidelines
Sample Submission Guidelines
 CNV detection in a validation group and a multicenter clinical group. (Dong, Z., et al., 2016)
CNV detection in a validation group and a multicenter clinical group. (Dong, Z., et al., 2016) An Overview of Japan's "Action Plan for Whole Genome Analysis of Cancer and Rare/Intractable Diseases" WGS Initiative (Yuki Katsuya, 2024)
An Overview of Japan's "Action Plan for Whole Genome Analysis of Cancer and Rare/Intractable Diseases" WGS Initiative (Yuki Katsuya, 2024) Workflow of method development and evaluation with low-pass genome sequencing for the paternity test. (Li, Keying, et al., 2023)
Workflow of method development and evaluation with low-pass genome sequencing for the paternity test. (Li, Keying, et al., 2023)