Since the inception of genome sequencing technology in the late 20th century, it has undergone rapid advancement and transformation. From the pioneering Sanger sequencing method to today's high-throughput sequencing technologies, scientists are now able to decipher the blueprint of life with unprecedented speed and precision. Within this evolutionary journey, low-pass whole-genome sequencing (LP-WGS) has emerged as a cost-effective genomic analysis approach that has gradually garnered attention.
As its name suggests, low-pass whole-genome sequencing involves sequencing the entire genome, albeit at a lower depth. Unlike high-throughput sequencing, LPWGS does not aim for deep coverage of every genomic region. Instead, it provides an overview of the genome through its reduced sequencing depth. This method is particularly suitable for large-scale population genetics studies, genome-wide association studies (GWAS), and certain types of genetic variation detection.
The concept of LPWGS can be traced back to the early stages of genome sequencing technology when technical limitations and high costs often restricted scientists to sequencing specific genes or gene fragments. With advancements in sequencing technology and a reduction in costs, whole-genome sequencing became attainable. Nonetheless, the expenses associated with comprehensive genome sequencing remain significant, especially when analyzing a large number of samples. LPWGS emerged as a means to balance research quality with cost efficiency.
Low-pass whole genome sequencing represents a cost-effective approach for genomic analysis, making it particularly suited for large-scale population genetics studies, GWAS, and the detection of genetic variations. This method provides a comprehensive overview of the entire genome without necessitating high-coverage depth, thus striking a balance between cost and research quality. LPWGS is capable of identifying copy number variations (CNVs) and single nucleotide polymorphisms (SNPs), and finds extensive applications in fields such as agriculture, ecology, and evolutionary biology. Although the amount of information yielded by LPWGS is not as exhaustive as that obtained from high-depth sequencing, advancements in sequencing technologies and decreasing costs are likely to enhance its significance in future genomic research.
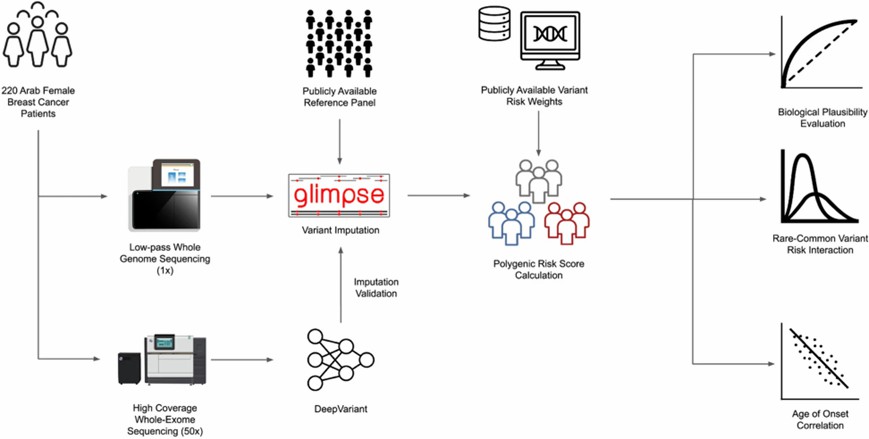 Low-pass whole genome sequencing (lpWGS) and high-coverage whole-exome sequencing (WES) were performed on blood samples collected from 220 indigenous Arab breast cancer patients. (Al-Jumaan, M., et al., 2023)
Low-pass whole genome sequencing (lpWGS) and high-coverage whole-exome sequencing (WES) were performed on blood samples collected from 220 indigenous Arab breast cancer patients. (Al-Jumaan, M., et al., 2023)
Definition of Low-Pass Whole Genome Sequencing
Low-pass whole genome sequencing represents a genomic sequencing technique characterized by analyzing the entirety of an individual's genome at a relatively shallow depth, typically achieving coverage below 10x. This method is specifically designed to quickly acquire a broad overview of an individual's genomic data while minimizing costs, avoiding the need for the extensive coverage and depth that defines traditional whole genome sequencing (WGS).
LPWGS offers several distinct advantages over conventional WGS. Firstly, regarding cost-efficiency, LPWGS demands significantly lower financial investment due to its reduced sequencing depth, thereby preserving valuable sequencing resources. Secondly, in terms of processing speed, LPWGS inherently accelerates the workflow because of the smaller volume of sequence data, facilitating more rapid data management and analysis. However, it is vital to acknowledge that the reduced sequencing depth in LPWGS may lead to a decrease in accuracy when detecting SNPs and minor insertions or deletions. Nonetheless, for purposes like large-scale population studies or preliminary genomic screenings, LPWGS remains a highly cost-effective alternative.
Comparison of Low-Pass Whole Genome Sequencing and Traditional Whole Genome Sequencing
| Comparison Metric |
Low-Pass Whole Genome Sequencing |
Traditional Whole Genome Sequencing (WGS) |
| Sequencing Depth |
Typically below 10x |
Higher |
| Cost |
Lower |
Higher |
| Processing Time |
Faster |
Slower |
| Data Volume |
Less |
More |
| Data Processing and Analysis Speed |
Faster |
Slower |
| Detection Accuracy |
May be lower than WGS |
Higher |
| Applicable Scenarios |
Large-scale population studies, preliminary screening |
Detailed analyses requiring high coverage and depth |
Advantages of Low-Pass Whole Genome Sequencing
As an emerging approach to DNA analysis, low-pass whole genome sequencing represents an economical method that balances data generation with practical utility. This technique achieves meaningful cost reductions through decreased sequencing depth while maintaining adequate genomic coverage for numerous applications.
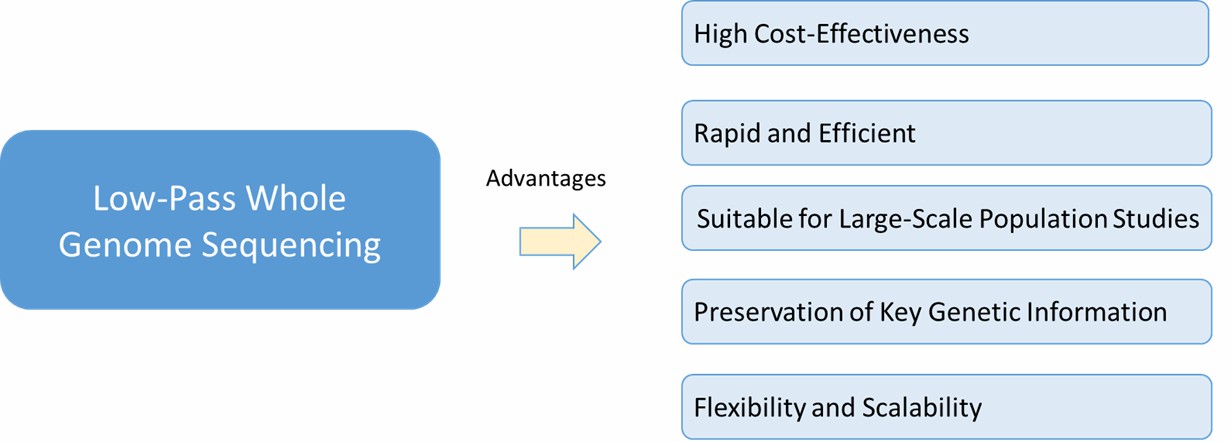 Some advantages of low-pass whole genome sequencing.
Some advantages of low-pass whole genome sequencing.
Key advantages of this methodology include:
- High Cost-Effectiveness: The reduced depth approach of LP-WGS substantially lowers operational costs when compared to conventional sequencing methods. By minimizing the number of required reactions, this technique optimizes resource utilization, enabling broader research scope within constrained financial parameters.
- Rapid and Efficient: Data generation and interpretation proceed more rapidly due to the streamlined nature of low-pass sequencing. The condensed dataset expedites computational analysis, proving especially valuable when time-sensitive results are crucial.
- Suitable for Large-Scale Population Studies: For large cohort analyses such as genome-wide association research, this methodology demonstrates particular utility. The approach enables comprehensive examination across numerous samples while retaining statistically meaningful data for variant detection.
- Preservation of Key Genetic Information: Although operating at reduced depth, the technique successfully identifies major genomic variations - encompassing polymorphisms, structural changes, and sequence modifications. This capability proves essential for understanding genotype-phenotype relationships.
- Flexibility and Scalability: The methodology serves effectively as a screening tool that can inform subsequent detailed analysis. Initial findings guide researchers in determining which genomic regions or sample sets warrant comprehensive sequencing, maximizing research efficiency.
This sequencing approach delivers a balanced solution for both research initiatives and clinical applications. As technology advances and expenses decrease further, low-pass sequencing techniques will likely assume greater significance in genomic investigation.
Services you may interested in
Case Studies of LP-WGS
Successful Cases of Low-Pass Whole-Genome Sequencing
Low-pass whole genome sequencing has demonstrated its effectiveness in various fields, particularly in clinical cytogenetics and agricultural genomics. In clinical cytogenetics, a study evaluated the performance of LP-WGS in detecting copy number variants (CNVs). The research analyzed 44 DNA samples, including both prenatal and postnatal cases, with a total of 55 chromosome imbalances. The chromosome imbalances ranged from 75 kb to 90.3 Mb, encompassing aneuploidies and cases of mosaicism. Remarkably, LP-WGS successfully detected all CNVs, showcasing its high consistency and robust performance.
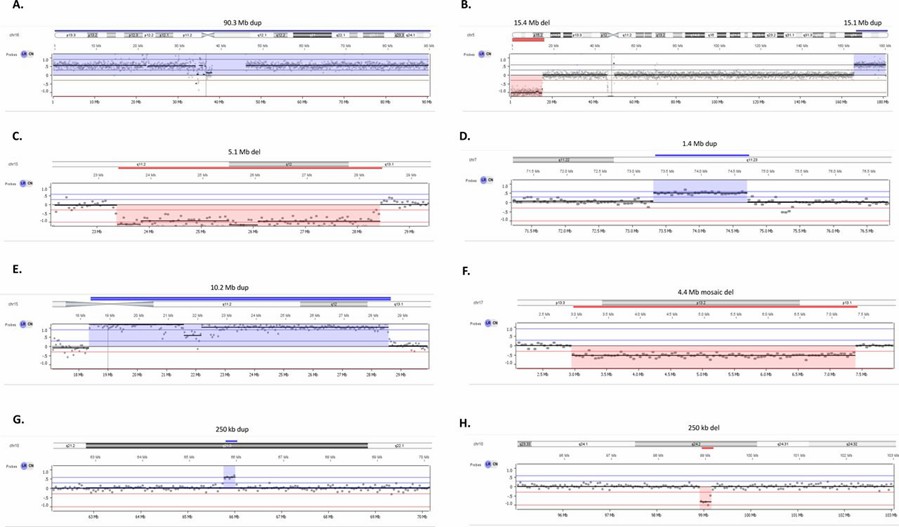 Copy number variants (CNVs) detected by low-pass whole genome sequencing (LP-WGS). (Mazzonetto, Patricia C., et al., 2024)
Copy number variants (CNVs) detected by low-pass whole genome sequencing (LP-WGS). (Mazzonetto, Patricia C., et al., 2024)
In agricultural genomics, LP-WGS has shown promise in revolutionizing breeding programs and enhancing food security. The technique offers a cost-effective alternative to traditional genotyping arrays, providing more accurate detection of genetic variation within the genomes of various species. For instance, researchers at the University of Queensland have successfully applied LP-WGS in both large-scale and smaller agricultural projects, demonstrating its wide applicability.
Case Analysis: Practical Effects of Technological Applications
The practical effects of LP-WGS are evident in its ability to provide high-quality genomic data at a lower cost and with shorter turnaround times compared to traditional sequencing methods. In a comparative study, LP-WGS was found to offer up to 99% accurate variant call detection and identification of SNPs, INDELs, and copy number variants3. This high accuracy, combined with its unbiased approach to variant detection, makes LP-WGS particularly valuable for GWAS, especially in underrepresented populations3.
In the field of agrigenomics, LP-WGS has demonstrated its potential to transform breeding practices. A study conducted by researchers at Northwest A&F University in China found that MGI's low-pass WGS solution provided farmers with better predictions at an affordable cost. This accessibility to advanced genomic tools could significantly impact agricultural practices, leading to more efficient and sustainable crop and livestock breeding programs.
Furthermore, LP-WGS has shown promise in clinical applications. A study published in Genome Biology demonstrated that LP-WGS, when combined with imputation techniques, could achieve comparable or even superior performance to genotyping arrays in trait mapping and polygenic risk score calculations. This finding suggests that LP-WGS could potentially replace genotyping arrays in many clinical and research settings, offering a more comprehensive view of genetic variation at a similar cost.
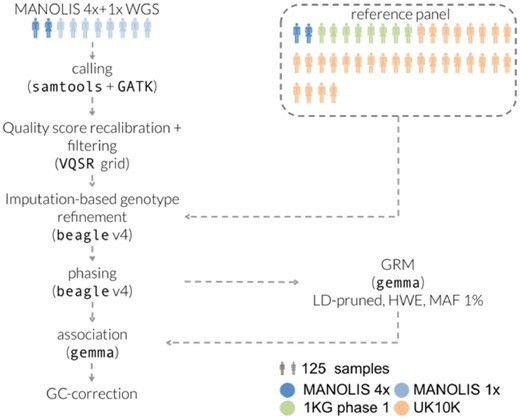 Processing pipeline for the MANOLIS 1× data. (Gilly, Arthur, et al., 2019)
Processing pipeline for the MANOLIS 1× data. (Gilly, Arthur, et al., 2019)
These case studies highlight the versatility and effectiveness of LP-WGS across different fields, from clinical diagnostics to agricultural breeding programs. As the technology continues to evolve and become more accessible, it is likely to play an increasingly important role in genomic research and applications.
Conclusion
As an advanced genomic sequencing technology, low-pass whole genome sequencing (LP-WGS) has demonstrated significant potential in recent years across clinical research, genetic disease diagnostics, and biodiversity analysis. This article aims to summarize the primary advantages of LP-WGS and to anticipate its future developments.
LP-WGS offers several key advantages:
Firstly, it is remarkably cost-effective. Compared to high-throughput sequencing, LP-WGS significantly reduces sequencing costs while maintaining a reasonable level of data quality. This reduction in cost makes whole genome sequencing accessible to a broader range of research and medical institutions, thereby facilitating the widespread adoption and application of sequencing technologies.
Secondly, data analysis is straightforward and rapid. The relatively smaller data output from LP-WGS translates to shorter data processing and analysis times. Researchers can obtain critical insights more swiftly, thus accelerating the pace of scientific discovery.
Furthermore, LP-WGS holds specific advantages in genetic disease diagnostics. It enables the rapid identification of genetic variants associated with hereditary diseases, providing clinicians with accurate diagnostic information to formulate more effective treatment strategies.
Additionally, LP-WGS shows immense promise in biodiversity analysis. By applying low-pass sequencing to various species, researchers can compare genomic differences, uncover evolutionary relationships, and assess phylogenetic affinities. Such insights provide robust support for biodiversity conservation and ecological research.
Looking ahead, the development of LP-WGS is poised to advance in several areas:
On one hand, with continuous improvements in sequencing technology, the accuracy and sensitivity of LP-WGS will see further enhancements, facilitating the precise detection of subtle genetic variations. This progress will provide more accurate data to support genetic disease diagnostics and drug development.
On the other hand, LP-WGS is expected to integrate deeply with cutting-edge technologies such as artificial intelligence and big data analytics, steering genomic sequencing toward a more intelligent and automated future. This evolution will significantly reduce costs, increase sequencing efficiency, and make genome sequencing more accessible and convenient.
In conclusion, LP-WGS stands out as a research focus in the field of genomics due to its cost-effectiveness, ease of data analysis, and unique advantages in genetic disease diagnostics and biodiversity analysis. As technological advancements continue and application fields expand, LP-WGS is expected to play an increasingly vital role, contributing more significantly to human health and biodiversity conservation.
References:
- Mazzonetto, Patricia C., et al. "Low-pass whole genome sequencing is a reliable and cost-effective approach for copy number variant analysis in the clinical setting." Annals of Human Genetics 88.2 (2024): 113-125. https://doi.org/10.1111/ahg.12532
- Gilly, Arthur, et al. "Very low-depth whole-genome sequencing in complex trait association studies." Bioinformatics 35.15 (2019): 2555-2561. https://doi.org/10.1093/bioinformatics/bty1032
- D'Agaro, E., Favaro, A., Matiussi, S. et al. Genomic selection in salmonids: new discoveries and future perspectives. Aquacult Int 29, 2259–2289 (2021). https://doi.org/10.1007/s10499-021-00747-w
- Zheng, Yu, et al. "Experience of low-pass whole-genome sequencing-based copy number variant analysis: a survey of chinese tertiary hospitals." Diagnostics 12.5 (2022): 1098. https://doi.org/10.3390/diagnostics12051098
- Al-Jumaan, M., Chu, H., Alsulaiman, A. et al. Interplay of Mendelian and polygenic risk factors in Arab breast cancer patients. Genome Med 15, 65 (2023). https://doi.org/10.1186/s13073-023-01220-4


 Sample Submission Guidelines
Sample Submission Guidelines
 Low-pass whole genome sequencing (lpWGS) and high-coverage whole-exome sequencing (WES) were performed on blood samples collected from 220 indigenous Arab breast cancer patients. (Al-Jumaan, M., et al., 2023)
Low-pass whole genome sequencing (lpWGS) and high-coverage whole-exome sequencing (WES) were performed on blood samples collected from 220 indigenous Arab breast cancer patients. (Al-Jumaan, M., et al., 2023) Some advantages of low-pass whole genome sequencing.
Some advantages of low-pass whole genome sequencing. Copy number variants (CNVs) detected by low-pass whole genome sequencing (LP-WGS). (Mazzonetto, Patricia C., et al., 2024)
Copy number variants (CNVs) detected by low-pass whole genome sequencing (LP-WGS). (Mazzonetto, Patricia C., et al., 2024) Processing pipeline for the MANOLIS 1× data. (Gilly, Arthur, et al., 2019)
Processing pipeline for the MANOLIS 1× data. (Gilly, Arthur, et al., 2019)