In the rapidly advancing field of genetics research, two approaches have emerged as particularly groundbreaking: Genome-Wide Association Studies (GWAS) and Whole Genome Sequencing (WGS). Although both techniques provide essential insights into the genetic foundations of traits and diseases, they differ markedly in their methodology, scale, and range of applications. This article seeks to explore these distinctions by analyzing the core principles behind each method, the extent of their utility, and the distinct benefits they offer. Additionally, it will examine the potential for combining these approaches, highlighting how their integration can enhance researchers' ability to push the limits of genetic discovery.
Technical Principles and Characteristics
GWAS is based on the principle of Linkage Disequilibrium (LD). It identifies potential disease or trait-associated genetic loci by analyzing the frequency of genetic variations in case and control groups within a large population. This method typically employs high-throughput genotyping technologies to detect and statistically analyze millions of single nucleotide polymorphism (SNP) sites across the genome, aiming to find variations that are significantly enriched in the case group. The advantage of this approach lies in its applicability to the study of complex traits and polygenic diseases, particularly in identifying common variants.
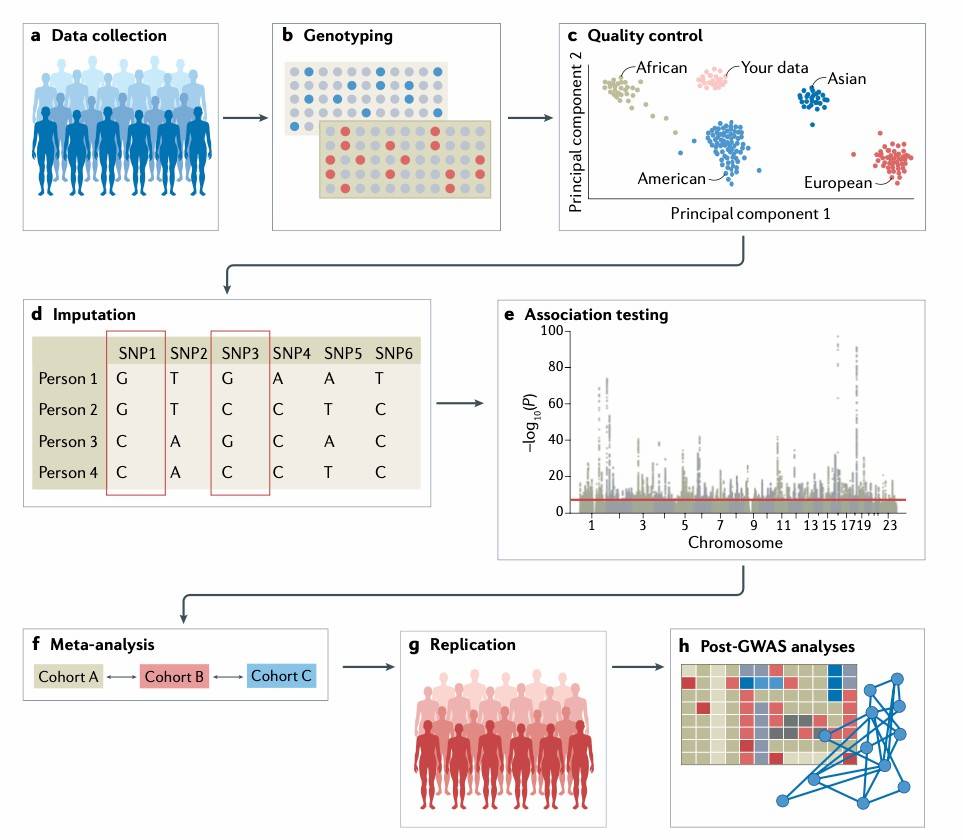 Figure 1.Overview of steps for conducting GWAS.(Uffelmann, et.al, 2021)
Figure 1.Overview of steps for conducting GWAS.(Uffelmann, et.al, 2021)
WGS is an all-encompassing genomic technique that employs high-throughput sequencing to map the complete genomic sequence of an individual, offering thorough genetic data. Unlike other methods, WGS can identify not only single nucleotide variants (SNVs) but also more intricate genomic alterations, including insertions, deletions, and structural changes. This technology plays a crucial role in detecting rare variants and de novo mutations linked to diseases, making it especially valuable in research on monogenic conditions and the advancement of personalized medicine.
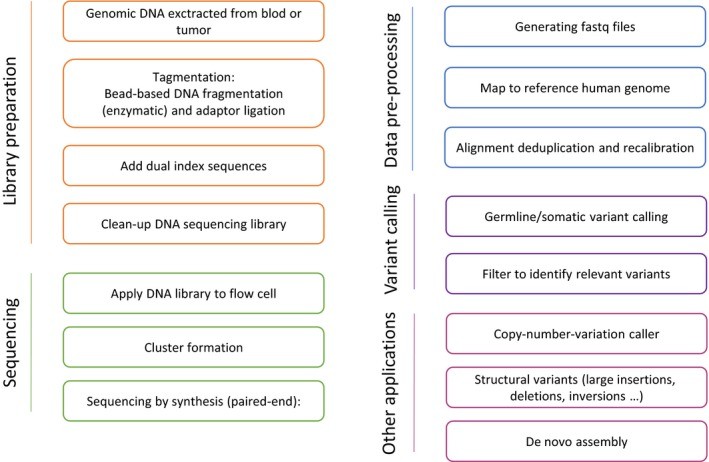 Figure 2.Whole-genome-sequencing workflow and bioinformatic pipeline.(Rossing, et.al, 2019)
Figure 2.Whole-genome-sequencing workflow and bioinformatic pipeline.(Rossing, et.al, 2019)
You may interested in
Resource
Research Scope and Depth
The focus of GWAS research is primarily on understanding the genetic underpinnings of common diseases and complex traits. By examining the frequency of genetic variations across large sample populations, it identifies widespread genetic factors linked to specific traits or diseases. Given its dependence on the statistical power derived from extensive sample sizes, GWAS is particularly well-suited for investigating complex genetic traits influenced by polygenic effects or gene-environment interactions, such as in conditions like diabetes, hypertension, and schizophrenia. However, the scope of GWAS is often confined to known SNP sites and common variants, limiting its ability to reveal the role of rare variants or structural changes. Consequently, GWAS is frequently employed as an initial approach for screening potential genetic loci associated with diseases, offering valuable insights for further functional validation and mechanistic exploration.
In contrast, WGS has a broader research scope and significantly enhanced depth of analysis. WGS not only reveals all known types of variations within an individual's genome but also uncovers de novo mutations, rare variants, and complex structural variants. As a result, WGS holds significant value in the study of rare diseases and monogenic disorders, particularly in uncovering the genetic basis of diseases with unknown etiology. Additionally, WGS is increasingly applied in personalized medicine and precision medicine, such as detecting key genomic variations related to drug responses or disease susceptibility, thereby providing personalized treatment strategies for patients.
Advantages and Limitations
Both GWAS and WGS offer distinct benefits and face specific limitations. GWAS is cost-effective and faster, making it ideal for large-scale studies focused on common diseases and complex traits. However, its capacity to detect rare variants and explore complex disease mechanisms is constrained. On the other hand, WGS provides complete genomic coverage, allowing for detailed analysis of rare variants and intricate structural variations, thus offering valuable insights for the study of rare diseases and personalized medicine. Despite these advantages, the high cost and the complexity of data analysis involved with WGS restrict its broader use.
| Comparison Dimension |
GWAS |
Whole Genome Sequencing (WGS) |
| Research Focus |
Common diseases and complex traits |
Rare diseases, monogenic disorders, personalized medicine |
| Technical Principle |
Based on linkage disequilibrium, analyzing SNP frequency |
Sequencing the entire genome to obtain comprehensive data |
| Advantages |
- Low cost |
- Comprehensive genome coverage |
| - Requires smaller sample sizes |
- Detects rare and de novo variants |
| - Fast analysis |
- Wide application range |
| Limitations |
- Limited ability to detect rare variants |
- High cost |
| - Limited insights into complex disease mechanisms |
- Complex data analysis |
| - Requires larger sample sizes |
| Data Types |
SNPs (Single Nucleotide Polymorphisms) |
SNPs, insertions/deletions, structural variants, de novo mutations |
| Suitable Research |
Initial association analysis of complex traits and polygenic diseases |
In-depth exploration of genetic mechanisms, rare disease causes, precision medicine |
| Cost |
Relatively low |
High |
| Data Analysis Demand |
Relatively simple, relies on standard statistical methods |
Large-scale data requiring advanced bioinformatics tools |
| Sample Size Demand |
Small to moderate |
Moderate to large |
Applications of GWAS and WGS
Applications of GWAS
GWAS as an efficient and cost-effective genetic research tool, is widely applied in areas such as the study of genetic factors in diseases, agricultural breeding, ecological and evolutionary research, and the genetic analysis of complex traits. It enables the identification of disease mechanisms through the analysis of population genetic variations, supports the genetic improvement of crops and livestock, and advances ecological adaptation and biological evolution research. Additionally, it provides in-depth analysis of the polygenic effects underlying complex traits, laying a critical foundation for precision medicine and the development of various fields.
GWAS studies have revealed that the APOE gene is associated with the risk of Alzheimer's disease onset. This association is particularly significant for the APOE ε4 allele, which has been shown to increase the likelihood of developing Alzheimer's disease (AD) substantially(Bertram, et.al, 2009).
Applications of WGS
WGS as a comprehensive technology for analyzing an individual's genetic information, is widely applied across various fields. It provides crucial support in the diagnosis of rare diseases by identifying pathogenic variants, aids in personalized cancer treatment by helping to develop precise therapeutic strategies, and offers scientific guidance in genetic counseling for assessing family disease risks and reproductive decisions. Additionally, WGS plays a vital role in infectious disease research, personalized health management, and studies in evolutionary biology and population genetics. With its comprehensiveness and precision, WGS has advanced the progress of precision medicine and life sciences.
In one case involving WGS, a patient diagnosed with a rare genetic disorder was found to carry mutations that made them eligible for a clinical trial of a novel gene therapy. This discovery not only opened up new treatment avenues but also provided hope for better management of their condition. The patient's participation in this trial was facilitated by the insights gained from WGS, showcasing its potential to influence clinical pathways positively(Turro, et.al, 2020).
Combined Application of GWAS and WGS
The combined application of these two technologies allows for complementary advantages in both breadth and depth. For example, the associated loci identified by GWAS can be further validated and functionally explored through WGS, while the high-precision sequence data generated by WGS can provide additional background support for GWAS, especially in the study of subtle variations and complex genetic patterns. This integration significantly enhances research efficiency and data interpretability.
A study conducted WGS and GWAS analysis on 1,972 PD patients and 2,478 healthy controls, with a replication study involving 8,209 PD patients and 9,454 healthy controls. The study identified multiple genetic variations associated with PD, including a new risk variant rs61204179, as well as four previously reported risk variants in European populations (NUCKS1/RAB29-rs11557080, SNCA-rs356182, FYN-rs997368, VPS13C-rs2251086) and three variants related to the LRRK2 coding region (R1628V, G2385R). The study also revealed genetic correlations between European and Chinese populations, with data showing that 63.6% of the PD genetic variants were significantly associated with the Chinese population (P < 0.05). Through polygenic risk score (PRS) analysis, the study demonstrated that individuals in the highest quartile for PRS had a 3.9-fold higher risk of developing PD compared to those in the lowest quartile (Pan et al., 2023).
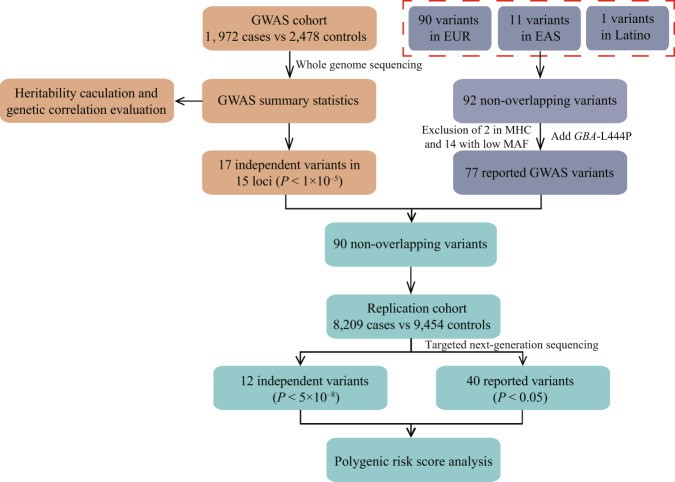 Figure 3.Workflow of unite GWAS and WGS.(Pan, et.al, 2023)
Figure 3.Workflow of unite GWAS and WGS.(Pan, et.al, 2023)
This combined study of Whole Genome Sequencing and GWAS reveals the genetic homogeneity and heterogeneity of Parkinson's disease across different ethnic populations. By identifying new genetic variants and comparing genetic correlations between populations, the study provides crucial data for a deeper understanding of the genetic mechanisms of Parkinson's disease. This research not only offers a theoretical foundation for identifying high-risk individuals in future precision medicine but also provides new perspectives and directions for genetic studies of Parkinson's disease.
Conclusion
GWAS and WGS each have their own advantages, providing powerful tools for genetic research. In practical studies, researchers can choose the appropriate technology based on factors such as research objectives, sample size, and budget. In the future, with ongoing advancements in technology, these two approaches will better serve human health and disease prevention.
References:
- Uffelmann, E., Huang, Q.Q., Munung, N.S. et al. Genome-wide association studies. Nat Rev Methods Primers 1, 59 (2021). https://doi.org/10.1038/s43586-021-00056-9
- Rossing, M., Sørensen, C. S., Ejlertsen, B., & Nielsen, F. C. (2019). Whole genome sequencing of breast cancer. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica, 127(5), 303–315. https://doi.org/10.1111/apm.12920
- Bertram, L., & Tanzi, R. E. (2009). Genome-wide association studies in Alzheimer's disease. Human molecular genetics, 18(R2), R137–R145. https://doi.org/10.1093/hmg/ddp406
- Turro, E., Astle, W. J., et.al. (2020). Whole-genome sequencing of patients with rare diseases in a national health system. Nature, 583(7814), 96–102. https://doi.org/10.1038/s41586-020-2434-2
- Pan, H., Liu, Z., et.al. (2023). Genome-wide association study using whole-genome sequencing identifies risk loci for Parkinson's disease in Chinese population. NPJ Parkinson's disease, 9(1), 22. https://doi.org/10.1038/s41531-023-00456-6


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1.Overview of steps for conducting GWAS.(Uffelmann, et.al, 2021)
Figure 1.Overview of steps for conducting GWAS.(Uffelmann, et.al, 2021) Figure 2.Whole-genome-sequencing workflow and bioinformatic pipeline.(Rossing, et.al, 2019)
Figure 2.Whole-genome-sequencing workflow and bioinformatic pipeline.(Rossing, et.al, 2019) Figure 3.Workflow of unite GWAS and WGS.(Pan, et.al, 2023)
Figure 3.Workflow of unite GWAS and WGS.(Pan, et.al, 2023)