Genome assembly is a core bioinformatics problem, which aims to reconstruct entire genomic sequences from short, segmented DNA reads. This is the basis of biological studies, enabling the investigation of genetic architecture, evolutionary dynamics, and functional genomics. The genome assembly process has been dramatically transformed with the advent of high-throughput sequencing technologies, moving away from laborious, low-throughput technologies like Sanger Sequencing to newer technology such as Illumina, PacBio and Oxford Nanopore. These technologies generate huge amounts of data, requiring computational algorithms to reconstruct genomes accurately and efficiently. In this article, we speak of importance of genome assembly in bioinformatics, its principles, computational strategies, challenges and applications via some real-world examples.
Bioinformatics addresses issues in Genome Assembly
While transformative, genome assembly has significant intrinsic challenges:
- Repeats: Eukaryotic genomes harbor large amounts of repetitive elements that complicate assembly. These regions are difficult to resolve and can lead to fragmented and inaccurate genome assemblies. Long-read sequencing instrumentation (e.g. PacBio HiFi and Nanopore) may emerge as a solution to this problem when it comes to crossing through repetitive regions.
- Heterozygosity: Differences between homologous chromosomes present in diploid and polyploid organisms blur the lines of distinction for haplotypes, posing assembly challenges. Recent phasing algorithms and haplotype-specific assemblers have significantly increased the phasing of heterozygous regions to reconstruct more complex genomes.
- Sequencing Dictionary Errors: While PacBio and Oxford Nanopore have long reads, they tend to make more mistakes. Reliable assemblies require effective error-correction mechanisms, such as consensus polishing tools (for example, Pilon and Racon).
- Data Megalomania: Large genomes produce an enormous amount of data that requires high computational resources for assembly, storage, and analysis. Managing these demands requires efficient algorithms and scalable infrastructure, for example cloud-based assembly pipelines.
Tackling these issues will require a suite of novel computational methods, rigorous algorithms, and context-dependent optimised workflows.
Service you may interested in
Resource
Bioinformatics Methods in Genome Assembly
Genome assembly methods reconstruct genomic sequences from millions or billions of short or long DNA reads.
De Novo Assembly
De novo assembly reconstructs genomes without the use of a reference genome, relying solely on the relationships shared between sequencing reads. This approach is essential to characterize novel organisms and to elucidate unique genomic characteristics.
- De Bruijn Graphs (DBG): DBG-based assemblers (like SPAdes and Velvet) break reads into smaller pieces called k-mers. These are used to build a graph with k-mers as nodes and overlaps as edges. This method shines at short-read assembly, making it a stellar choice for both microbial genomes and small eukaryotes.
- Overlap-Layout-Consensus (OLC): OLC algorithms, with implementations such as Canu and Flye, find overlaps between long reads, create layouts and generate consensus sequences. This approach is particularly suited to overcoming repeats and structurally complex regions of genomes and delivers much more contiguous assemblies. Right now, the OLC assemblers have been updated recently to add error tolerant methods for the noisy long-read data improving the applicability even further.
Application of Bioinformatics in Genome Assembly
- Single-cell Genomics: Single-cell genome assembly has the promise to reveal genetic heterogeneity at an unprecedented level of resolution. In tumor research, this is especially relevant in that the heterogeneity of tumors is critical for their behavior in growth and therapy resistance. The microbiome studies also rely heavily on single-cell methods to study microbial diversity and symbiotic relationships.
- Multi-omics Integration: There is a growing trend to integrate genome assembly with other omics data, such as transcriptomics, proteomics, and epigenomics. This integrated strategy gives a complete view of gene function, regulation, and interactions, linking the static nature of genomic sequences and dynamic biological processes.
- AI-Powered Assembly Pipelines: Several groups are leveraging machine learning and artificial intelligence to impact genome assembly. These technologies improve error-correction, repeat-resolution and structural variant detection, while also simplifying the computational workflow. Machine learning can make predictions about optimal assembly parameters, leading to improvements in performance while reducing the amount of calculation required.
- Open-Access Genomic Resources: International efforts such as the Earth BioGenome Project seek to sequence and assemble the genomes of all eukaryotic species. These initiatives are democratizing genomic data - enabling researchers around the world to study biodiversity and tackle urgent ecological and societal issues - by standardizing workflows and establishing open-access databases.
- Description: Precision medicine requires high-quality genome assemblies to identify rare genetic variants causing diseases. As sequencing costs continue to plummet while assembly tools improve, clinical genomics will become the norm and offer individualized insights into diagnostic and treatment strategies.
Bioinformatics significance in Genome Assembly
Genome assembly is at the center of modern genomics but forms the basis to retrieve biological information of interest from sequencing data. Its importance is evident from its extensive applications and the solutions it provides to basic scientific questions.
Genome assembly opens up myriad opportunities for discovery and innovation:
- Gene Identification and Annotation: Genome assembly is fundamental for the identification of coding and non-coding regions, regulatory elements, and structural features. It is this foundational knowledge that underpins functional genomics, allowing scientists to investigate the molecular basis of biological phenomena and pathology. Assembled genomes also serve as references for transcriptome mapping, providing an understanding of tissue-specific gene expression and alternative splicing patterns.
- Comparative and Evolutionary Genomics: When scientists have high-quality assemblies, they can compare their genomes to those of other species and find genes that are conserved and ones that are unique adaptations. Such comparisons reveal evolutionary lineages and speciation events, as well as the genetic basis of phenotypic diversity. As this approach has been applied to comparative analysis of human and primate genomes, insights have emerged into key genomic regions controlling traits such as cognitive development and immunological variation.
- Human Health: Assemblies of human and pathogen genomes are essential to characterize disease-associated mutations, track the evolution of infectious agents and to develop vaccines. As an example, SARS-CoV-2 genome assemblies have been critical for the design of diagnostics and therapeutics targeting COVID-19. In a similar vein, cancer genomics depends on assembling tumor genomes to discover driver mutations and therapeutic targets.
- Agricultural applications and breeding: Genome assemblies of crops help identify genes for yield, disease resistance, and stress tolerance. Such information facilitates precision breeding, thereby speeding up creating resilient crop varieties to tackle the challenges of global. Rice and maize genome assembly, for instance, has exposed important genes linked with drought tolerance and pest resistance thereby enhancing agricultural practices all through.
- Environmental Genomics: Reconstructing microbial genomes from various environments to understand their ecological roles, metabolic capabilities, and interactions. This knowledge contributes to our understanding of biodiversity and biogeochemical cycles and can inform conservation and environmental sustainability efforts. A notable example is the reconstruction of metagenome-assembled genomes (MAGs) from marine habitats which have revealed novel aspects of microbial mediation of carbon cycling and nitrogen fixation.
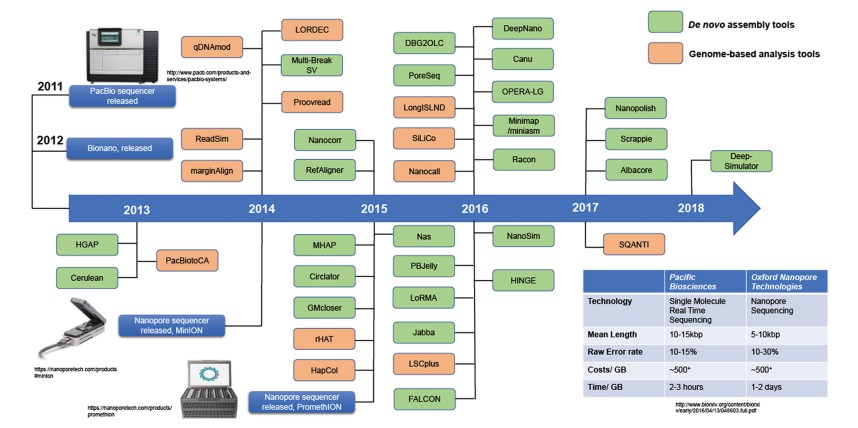 Milestones in TGS analysis software development (Wee, Y. et al 2019).
Milestones in TGS analysis software development (Wee, Y. et al 2019).
Case Study: Wheat Genome Assembly
Background
Encompassing around 17 Gb in size, the wheat genome is the most complex of all genetically significant crops, displaying hexaploidy - containing three homologous subgenomes. Assembly was particularly difficult due to repetitive regions and high heterozygosity. Improving crops for resilience under climate change and drones, or more productive under climate change, is indispensable and so, understanding the genome of wheat is critical to achieving global food security.
Methods
- Sequencing technologies: in the two most recent projects, a hybrid sequencing approach was applied, combining short reads (Illumina) and long reads (PacBio, Oxford Nanopore) for both accuracy and continuity. Scaffold construction was then refined further by optical mapping and Hi-C data.
- Assembly tools: Canu (long reads), SPAdes (short reads) and MaSuRCA (hybrid datasets) Hi-C based assembly tools were utilized for scaffolding and gap filling.
- Validation: Assembly quality for all genomes was assessed using metrics like BUSCO completeness scores and QUAST contiguity statistics, whereas comparative analyses with other closely related species validated both structural and functional accuracy.
Results
The final assembly achieved a scaffold N50 of >10 Mb and successfully resolved complex repetitive regions and structural variants.Identification of key genes related to disease resistance (rust) and abiotic stress tolerance These insights have been utilized by marker-assisted selection programs to speed up the breeding process.
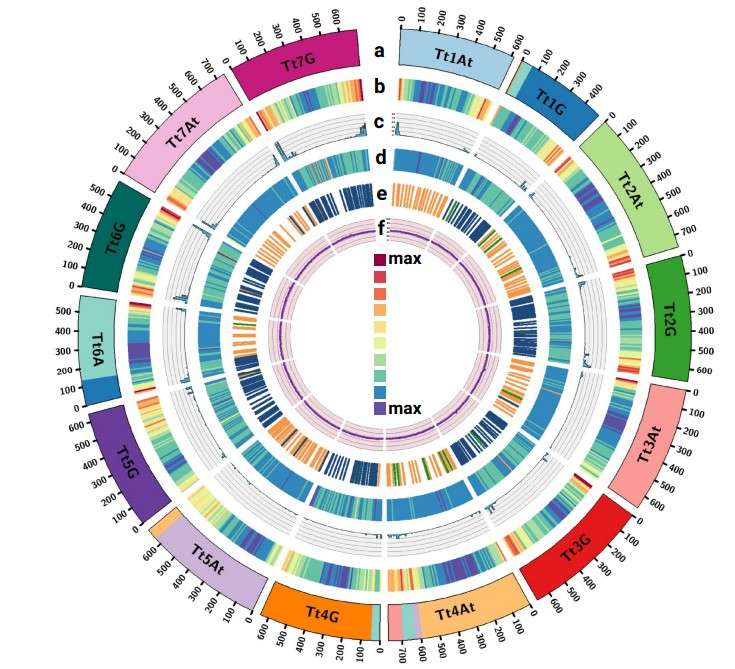 Features of the chromosome-scale assembly (Grewal, S. et al 2024).
Features of the chromosome-scale assembly (Grewal, S. et al 2024).
Conclusion
Genome assembly is a core bioinformatics task, enabling the fundamental study of the genetic blueprint of life. By aiding basic discoveries in evolution and ecology and allowing for medical and agricultural breakthroughs, genome assembly has revolutionized our ability to decode and understand complex genomes. Pairs with relatively long reads, which make it possible to construct longer overlaps, small genomes with relatively short reads, and more powerful assembly algorithms have caused the genome assembly landscape to evolve remarkably in recent years.
These expansions in the capabilities of genome assembly that have been made possible by recent advances in ultra-long-read sequencing, single-cell genomics, and AI-based methods will continue to drive innovations in this field. Not only will these improvements increase the accuracy and speed of assemblies, but they will also enable new avenues in studying biological diversity, complexity, and function. Genome assembly will continue to be at the leading edge of scientific research as this field grows and matures, paving the way for revolutionary measures across all fields and the future of genomics.
References:
- Wee, Y., Bhyan, S. B., Liu, Y.,et.al. (2019). The bioinformatics tools for the genome assembly and analysis based on third-generation sequencing. Briefings in functional genomics, 18(1), 1–12. https://doi.org/10.1093/bfgp/ely037
- Grewal, S., Yang, C. Y., Scholefield, D., et.al. (2024). Chromosome-scale genome assembly of bread wheat's wild relative Triticum timopheevii. Scientific data, 11(1), 420. https://doi.org/10.1038/s41597-024-03260-w


 Sample Submission Guidelines
Sample Submission Guidelines
 Milestones in TGS analysis software development (Wee, Y. et al 2019).
Milestones in TGS analysis software development (Wee, Y. et al 2019). Features of the chromosome-scale assembly (Grewal, S. et al 2024).
Features of the chromosome-scale assembly (Grewal, S. et al 2024).