The genetic framework of botanical species can be comprehensively understood through the meticulous analysis of chloroplast genetic material. As fundamental organelles responsible for converting solar energy into chemical energy, chloroplasts represent critical components within plant cellular structures. Scientific investigations into chloroplast genomic sequences provide profound insights into evolutionary mechanisms, genetic variation, and the fundamental characteristics of plant populations. This molecular approach holds transformative potential across multiple research domains, including evolutionary biology, ecological preservation efforts, and advanced agricultural research methodologies.This article delves into the core concepts, methodologies, and practical uses of cpDNA sequencing, offering a detailed resource for both researchers and enthusiasts.
What is Chloroplast DNA (cpDNA)?
Genetic material contained within photosynthetic organelles represents a sophisticated molecular system distinct from nuclear genomic structures. Chloroplast DNA demonstrates unique inheritance patterns, characterized by circular configuration and predominantly maternal transmission across numerous botanical species. Unlike nuclear genetic elements transferred through biparental mechanisms, this specialized genomic configuration provides distinct insights into plant evolutionary mechanisms.
Regarding genomic dimensions, chloroplast genetic material exhibits remarkable consistency in higher botanical classifications. Typically measuring between 120 and 160 kilobase pairs, these molecular structures contrast with algal genomic arrangements that display more substantial dimensional variability. The comprehensive genetic landscape encompasses four primary structural domains: large single-copy regions, small single-copy segments, and two symmetrically positioned inverted repetitive sequences.
The intricate architectural design of chloroplast genetic elements, featuring reciprocally oriented identical sequence segments, enables sophisticated scientific investigations into organelle function and evolutionary dynamics. This complex molecular organization provides researchers with critical frameworks for understanding botanical genetic progression and cellular metabolic processes.
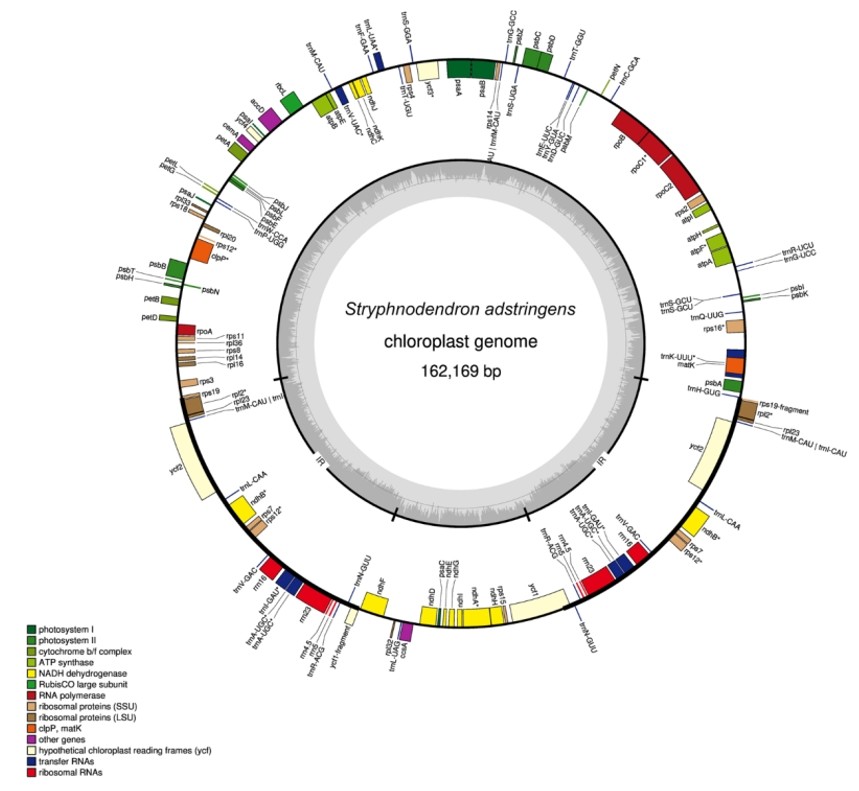 Figure 1. Gene map of the S. adstringens chloroplast genome. (Souza, U.J.B.d., et.al, 2019)
Figure 1. Gene map of the S. adstringens chloroplast genome. (Souza, U.J.B.d., et.al, 2019)
The cpDNA structure in algae is more complex, with inverted repeat sequences (IR) commonly absent. In species of algae that retain IRs, these sequences often exhibit signs of shortening and degradation. Additionally, the cpDNA of algae has higher coding capacity, typically containing more genes than that of higher plants.
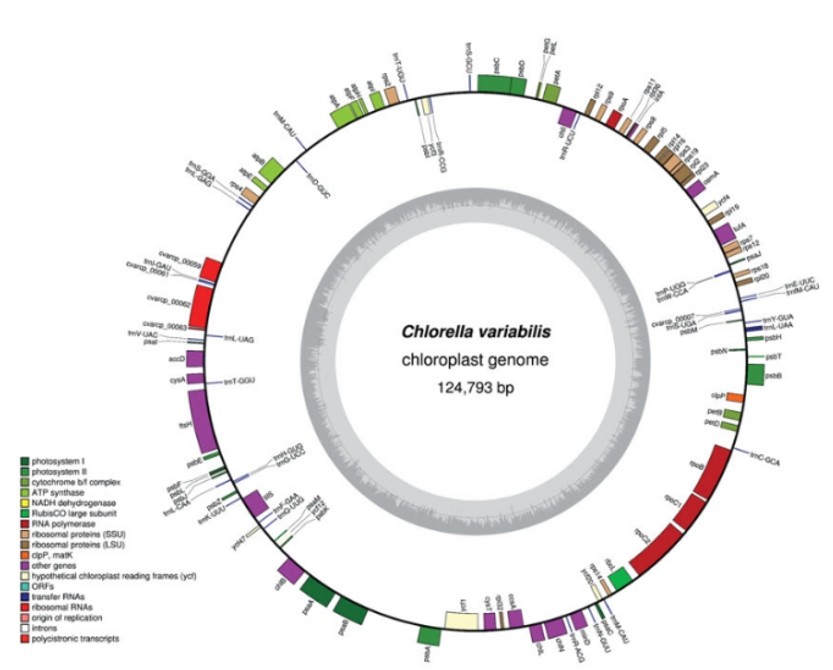 Figure 2. Gene maps of the chloroplast genomes of C. variabilis. (Orsini, M., et.al, 2016)
Figure 2. Gene maps of the chloroplast genomes of C. variabilis. (Orsini, M., et.al, 2016)
CpDNA plays a key role in plant cell functions, such as energy production and synthesis of essential compounds. This makes it an excellent target for genetic studies, including those aimed at understanding plant evolution, species identification, and biodiversity conservation.
Principles of Chloroplast DNA Sequencing
Molecular genetic investigations of chloroplast genomic material involve sophisticated methodological approaches for extracting and analyzing botanical genetic components. The scientific procedure typically initiates with precise isolation of genetic material from specific plant tissue samples, utilizing targeted molecular amplification techniques to investigate genomic regions of particular research interest.
Researchers exploring phylogenetic relationships among botanical species—such as those within the Vaccinium genus—employ strategic genetic extraction protocols to compare molecular sequences across different taxonomic specimens. Contemporary genomic analysis leverages advanced technological platforms, including high-resolution sequencing methodologies like Illumina , Nanopore and PacBio systems, which enable comprehensive molecular mapping with exceptional precision(Singh et al., 2021).Technological innovations in Next-Generation Sequencing (NGS) have revolutionized botanical genetic research, facilitating rapid and comprehensive examinations of extensive chloroplast genomic segments.
The fundamental objective of chloroplast genetic sequencing centers on deciphering the intricate nucleotide arrangements that constitute plant genomic architectures. Investigators systematically identify the specific sequential configuration of molecular building blocks(A, T, C, G) to unravel complex genetic relationships and evolutionary trajectories.
Common Methods of Chloroplast DNA Sequencing
Sanger Sequencing
Sanger sequencing represents the conventional approach to DNA sequencing. It is known for its high accuracy and remains a preferred choice for smaller-scale projects or situations where precision is paramount. Nevertheless, this method has limitations in throughput and tends to be more time-intensive compared to more modern techniques.
Next-Generation Sequencing (NGS)
Platforms for Next-Generation Sequencing, such as Illumina and PacBio, provide high-throughput capabilities by allowing the simultaneous sequencing of millions of DNA fragments. This advancement considerably decreases both the time and cost involved, making it particularly suitable for extensive projects, including cpDNA sequencing across a variety of plant species. NGS technologies enable researchers to sequence entire genomes or specific regions of cpDNA in one operation, yielding more detailed data at a fraction of the expense associated with traditional methods.
Whole Genome Shotgun Sequencing
Whole genome shotgun sequencing (WGS) is a methodology that involves randomly breaking DNA into small segments, which are then sequenced independently. This approach is particularly effective for acquiring cpDNA information from complex genomic libraries, as it offers a more comprehensive view of the chloroplast genome in a single procedure.
Data Analysis of Chloroplast DNA Sequencing
Data analysis of chloroplast DNA sequencing typically involves several key steps to ensure high-quality results and reliable interpretations.
The first step is quality control of raw sequencing data, which involves removing low-quality reads and adapter sequences. This ensures that downstream analyses are not compromised by technical errors. Next, clean reads are aligned to a reference chloroplast genome or de novo assembled to reconstruct the cpDNA. Advanced bioinformatics tools such as Bowtie2, SPAdes, and GetOrganelle are widely used in this process.
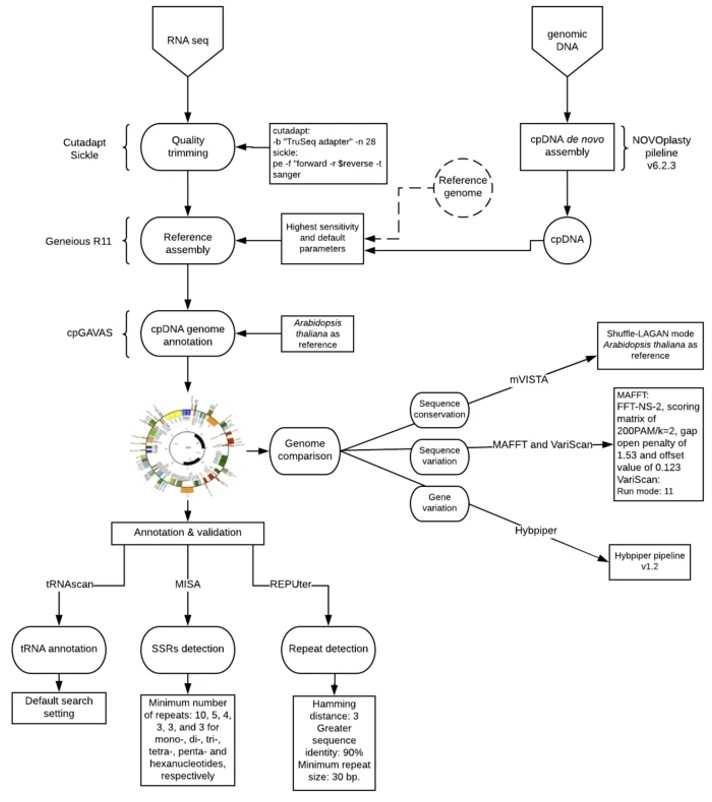 Figure 3. A flow chart depicting the bioinformatics analyses to assembly cp genomes. (Osuna-Mascaró, C., et.al, 2018)
Figure 3. A flow chart depicting the bioinformatics analyses to assembly cp genomes. (Osuna-Mascaró, C., et.al, 2018)
Detecting genetic variants, such as SNPs and indels, is a fundamental aspect of examining genetic diversity within or among species. Annotating chloroplast genomes allows researchers to pinpoint coding and non-coding regions, including genes crucial for photosynthesis and other vital metabolic functions. Phylogenetic relationships are commonly inferred through specialized tools like MEGA or RAxML, enabling detailed evolutionary studies.
Chloroplast DNA sequencing serves as a powerful tool for investigating plant genetics, facilitating species identification, promoting biodiversity conservation, and shedding light on how plants adapt to varying environmental conditions.
Applications of Chloroplast DNA Sequencing
Chloroplast DNA sequencing has various applications across multiple research fields:
Phylogenetics
Analyzing chloroplast DNA (cpDNA) allows researchers to explore the evolutionary connections among plant species. Phylogenetic analyses are instrumental in tracing the origins of plant lineages, reconstructing their evolutionary histories, and examining the diversification of plant families. For instance, a study focusing on the Chaenomeles genus utilized chloroplast genome data to uncover phylogenetic relationships, demonstrating that Chaenomeles forms a monophyletic group closely associated with Docynia and Malus species. This research offered valuable insights into the evolutionary pathways of these plants and shed light on their diversification within the Rosaceae family (Sun, J., et al., 2020).
Conservation Biology
Chloroplast DNA sequencing is a powerful tool in conservation biology. By assessing genetic diversity within plant populations, especially endangered species, researchers can identify genetically unique populations and inform conservation strategies. For instance, researchers studying endangered plant species like Pseudotsuga menziesii (Douglas fir) have utilized cpDNA sequencing to assess genetic diversity within populations. By identifying genetically unique populations, they can develop targeted conservation strategies that preserve these vital genetic resources and enhance the resilience of the species against environmental changes(De-la-Peña, C.,et.al ,2022).
Agriculture
Molecular genetic analysis of chloroplast DNA has emerged as a sophisticated strategy for agricultural advancement and crop enhancement. Comprehensive genomic investigations of cultivated and wild plant populations facilitate the development of innovative agricultural varieties with enhanced physiological characteristics. Researchers can strategically identify and integrate genetic markers associated with critical traits like pathogen resistance, productivity optimization, and environmental adaptability.
The Vaccinium genus represents a compelling illustration of this scientific approach. Systematic genomic sequencing of blueberry-related species provides researchers with nuanced insights into genetic variability and potential breeding strategies. By examining chloroplast genetic structures, plant scientists can develop more robust and productive cultivars that address emerging agricultural challenges.(Fahrenkrog, A.M., et.al, 2022).
Additionally, innovative techniques like modifying chloroplast genes without altering nuclear DNA have been proposed to create crops that are more resilient to climate change while maintaining consumer acceptance.
Ecology and Plant Ecology
CpDNA sequencing helps in understanding plant adaptation to various environmental conditions, enabling more efficient ecological conservation and management.For example, studies on the chloroplast genomes of plants in different habitats have revealed how specific genetic variations contribute to their adaptability in extreme environments, such as arid deserts or high-altitude regions. This information can guide conservation efforts by identifying which genetic traits are crucial for survival in changing climates(Liu, D., et.al, 2019).
Conclusion
Chloroplast DNA sequencing is a powerful tool in plant genetics research, helping scientists uncover the genetic makeup of plants and their evolutionary relationships. However, achieving accurate and comprehensive sequencing results requires specialized knowledge, advanced technologies, and high-quality services.
At CD Genomics, we offer state-of-the-art chloroplast DNA sequencing services using cutting-edge technologies like Next-Generation Sequencing (NGS). Our services are tailored to meet the needs of researchers in phylogenetics, conservation biology, and agriculture, ensuring high-quality results at competitive prices.
References:
- Souza, U.J.B.d., Nunes, R., Targueta, C.P. et al. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae - Caesalpinioideae): comparative analysis with related Mimosoid species. Sci Rep 9, 14206 (2019). https://doi.org/10.1038/s41598-019-50620-3
- Orsini, M., Costelli, C., Malavasi, V., Cusano, R., Concas, A., Angius, A., & Cao, G. (2016). Complete sequence and characterization of mitochondrial and chloroplast genome of Chlorella variabilis NC64A. Mitochondrial DNA. Part A, DNA mapping, sequencing, and analysis, 27(5), 3128–3130. https://doi.org/10.3109/19401736.2015.1007297
- Singh, N. V., Patil, P. G., Sowjanya, R. P., et.al. (2021). Chloroplast Genome Sequencing, Comparative Analysis, and Discovery of Unique Cytoplasmic Variants in Pomegranate (Punica granatum L.). Frontiers in genetics, 12, 704075. https://doi.org/10.3389/fgene.2021.704075
- Osuna-Mascaró, C., Rubio de Casas, R. & Perfectti, F. Comparative assessment shows the reliability of chloroplast genome assembly using RNA-seq. Sci Rep 8, 17404 (2018). https://doi.org/10.1038/s41598-018-35654-3
- Sun, J., Wang, Y., Liu, Y. et al. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep 10, 11466 (2020). https://doi.org/10.1038/s41598-020-67943-1
- De-la-Peña, C., León, P., & Sharkey, T. D. (2022). Editorial: Chloroplast Biotechnology for Crop Improvement. Frontiers in plant science, 13, 848034. https://doi.org/10.3389/fpls.2022.848034
- Fahrenkrog, A.M., Matsumoto, G.O., Toth, K. et al. Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops. Sci Rep 12, 21600 (2022). https://doi.org/10.1038/s41598-022-25434-5
- Liu, D., Cui, Y., Li, S., Bai, G., Li, Q., et.al.. (2019). A New Chloroplast DNA Extraction Protocol Significantly Improves the Chloroplast Genome Sequence Quality of Foxtail Millet (Setaria italica (L.) P. Beauv.). Scientific reports, 9(1), 16227. https://doi.org/10.1038/s41598-019-52786-2


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Gene map of the S. adstringens chloroplast genome. (Souza, U.J.B.d., et.al, 2019)
Figure 1. Gene map of the S. adstringens chloroplast genome. (Souza, U.J.B.d., et.al, 2019) Figure 2. Gene maps of the chloroplast genomes of C. variabilis. (Orsini, M., et.al, 2016)
Figure 2. Gene maps of the chloroplast genomes of C. variabilis. (Orsini, M., et.al, 2016) Figure 3. A flow chart depicting the bioinformatics analyses to assembly cp genomes. (Osuna-Mascaró, C., et.al, 2018)
Figure 3. A flow chart depicting the bioinformatics analyses to assembly cp genomes. (Osuna-Mascaró, C., et.al, 2018)