What is the ChIP-on- chip Method
1. Technical Overview
Modern genomic research employs various sophisticated analytical methods, among which the combination of chromatin immunoprecipitation with microarray analysis stands prominent. This dual-technology approach, termed ChIP-on-chip (also known as ChIP-chip), enables researchers to examine protein interactions with DNA across entire genomes. Notably, the methodology's name reflects its hybrid nature: chromatin immunoprecipitation (ChIP) paired with microarray analysis (chip).
2. Fundamental Purpose
This method helps reveal important details about how genes are regulated. It maps where specific proteins bind to DNA. By combining immunoprecipitation and microarray technology, scientists can see where proteins bind to DNA in the genome. This provides new insights into gene regulation.
3. Objectives and Applications
The primary objective of ChIP-on-chip (Chromatin Immunoprecipitation integrated with microarray technology) is to ascertain the binding sites of specific proteins within the genome, notably those of transcription factors and histone modifications involved in gene expression regulation. This technique finds extensive applications in the domains of gene expression regulation, chromatin structure analysis, and epigenetic research.
Common applications include:
- Transcription Factor Binding Site Analysis: Identifying the specific genomic regions where transcription factors bind, shedding light on their regulatory roles in gene expression.
- Histone Modification Analysis: Examining the distribution of various histone modifications, such as methylation and acetylation, within chromatin to explore their impact on gene activity.
- Epigenetic Studies: Investigating how epigenetic marks like DNA methylation and histone modifications influence gene expression, thus affecting cellular function and fate.
- Disease Mechanism Research: Unraveling molecular mechanisms of disease by studying interactions between disease-related proteins and the genome.
4. Comparative Benefits
When evaluated against traditional methods like ChIP-qPCR, this integrated approach delivers broader genomic insights. While ChIP-sequencing technologies offer higher resolution, ChIP-on-chip is still useful for many studies, especially when considering available resources and the specific needs of the experiment.
5. Research Impact
Through comprehensive examination of protein-DNA interactions, this methodology continues advancing our understanding of genomic regulation. Its applications span fundamental research through clinical investigations, contributing significantly to molecular biology knowledge.
Service you may intersted in
For more details, please refer to the following articles:
Process of ChIP-on-chip Technology
Investigating genome-wide protein-DNA binding patterns requires sophisticated experimental approaches. Through combining immunoprecipitation techniques with microarray analysis, researchers can systematically map molecular interactions across genomic regions. Success depends on executing multiple critical experimental phases with precision.
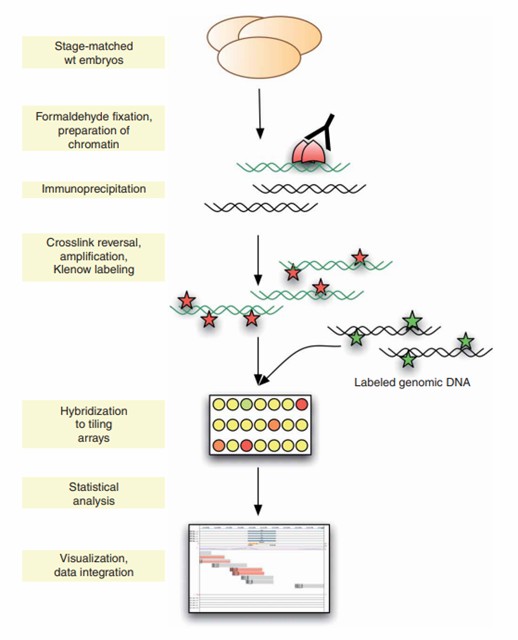 Schematical overview of ChIP-on-chip experiments. (Sandmann, T., et al., 2006)
Schematical overview of ChIP-on-chip experiments. (Sandmann, T., et al., 2006)
Detailed Experimental Stages
Phase 1: Initial Sample Preparation and Crosslinking
Preserving molecular interactions begins with stabilization:
- Apply formaldehyde treatment to create stable protein-DNA bonds
- Neutralize excess crosslinking agent using glycine solution
- Perform sequential buffer washes to eliminate contaminants
- Disrupt cellular structures using specialized lysis solutions containing detergents
- Maintain protein stability throughout membrane disruption
Phase 2: DNA Fragment Generation
Optimal analysis requires precise chromatin processing:
- Generate DNA segments ranging from 200-500 nucleotides
- Utilize either physical disruption via sonic waves
- Alternatively, employ enzymatic processing with MNase
- Monitor fragment distribution for size consistency
- Verify fragmentation efficiency through gel analysis
Phase 3: Selective Protein-DNA Complex Isolation
Target specific molecular complexes through:
- Selection using validated antibodies against proteins
- Capture complexes via protein A/G conjugated matrices
- Remove non-specific materials through centrifugation
- Implement stringent washing procedures
- Maintain complex integrity throughout isolation
Phase 4: Complex Purification
Optimize sample purity through sequential processing:
- Begin with mild buffer conditions
- Progress to increasingly stringent wash solutions
- Apply controlled temperature elevation
- Maintain appropriate ionic conditions
- Release DNA through crosslink reversal
Phase 5: Microarray Analysis
Final analytical procedures include:
- Purify isolated DNA fragments
- Incorporate fluorescent markers
- Perform controlled probe hybridization
- Scan arrays for signal detection
- Process data using specialized software
Technical Considerations
Quality Control Measures
- Validate antibody specificity
- Monitor fragment size distribution
- Assess enrichment efficiency
- Verify signal-to-noise ratios
- Implement appropriate controls
Critical Parameters
- Maintain precise timing during crosslinking
- Control sonication parameters
- Optimize wash conditions
- Ensure proper temperature control
- Verify probe specificity
In conclusion, ChIP-on-chip technology empowers researchers to comprehensively understand protein-DNA interactions, elucidating their roles in gene expression regulation, chromatin remodeling, and disease pathogenesis.
What is the ChIP Method
ChIP is a foundational experimental technique widely employed in the realm of epigenetics. It serves as an essential tool for elucidating the interactions between specific proteins and DNA, thereby deepening our understanding of the regulatory mechanisms governing gene expression.
Principle of ChIP
ChIP operates on the principle of stabilizing protein-nucleic acid interactions within living cells by inducing covalent bonds through formaldehyde treatment. In the cellular environment, transcription factors (TFs) often bind to promoter regions, resulting in close proximity or direct contact between these molecules. Formaldehyde facilitates the fixation of these interactions by forming covalent bonds between the proteins and DNA. Following this stabilization, the chromatin is fragmented into smaller, defined segments via sonication or enzymatic digestion. The resulting protein-DNA complexes are enriched using antigen-antibody specificity, allowing for the selective precipitation of DNA fragments bound to the target protein. These fragments are subsequently purified and analyzed to yield pivotal insights into protein-DNA interactions. Techniques such as quantitative PCR (qPCR) or next-generation sequencing are employed to identify novel DNA sequences interacting with the target proteins.
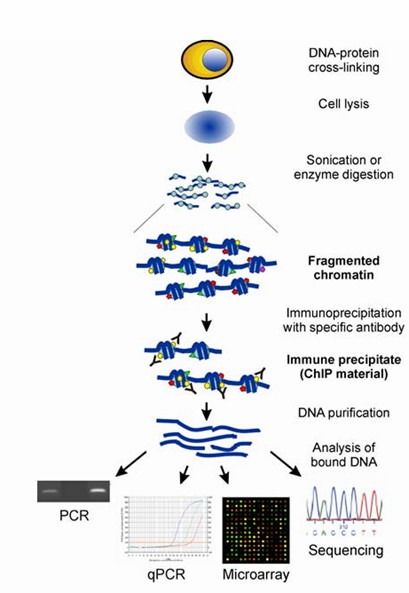 The chromatin immunoprecipitation (ChIP) assay and various methods of analysis. (Collas, et al., 2010)
The chromatin immunoprecipitation (ChIP) assay and various methods of analysis. (Collas, et al., 2010)
Building on this concept, ChIP and its derivative techniques—ChIP-chip, ChIP-seq, ChIP-PCR, and ChIP-qPCR—provide powerful methodologies for the exploration of protein-DNA interactions.
When is ChIP used?
- Transcription Factors: ChIP is instrumental in charting the binding sites and sequences of specific transcription factors across the genome. This capability allows for the identification of cis-regulatory elements of downstream genes, thereby enhancing our comprehension of the signaling pathways mediated by these transcription factors.
- Transcription Mechanisms: ChIP is utilized to delineate the binding sites of RNA Polymerase II and other transcriptional constituents, unveiling promoter and enhancer sequences and potentially leading to the discovery of novel transcriptional regulatory mechanisms.
- Histone Modifications: In the study of histone modifications, ChIP plays a critical role in uncovering and characterizing the histone code. It enables the detection of specific histone modifications and their regulatory roles in gene expression.
Through such applications, ChIP emerges as a formidable tool in the study of protein-DNA interactions, offering profound insights into gene regulation, chromatin remodeling, and the molecular underpinnings of diverse biological processes.
What is the Difference Between ChIP-seq and ChIP-on-chip
ChIP-seq (Chromatin Immunoprecipitation combined with high-throughput sequencing) and ChIP-on-chip are both essential methodologies for investigating protein-DNA interactions. Although they serve the same general purpose, they differ substantially in their experimental frameworks, data analysis techniques, and range of applications.
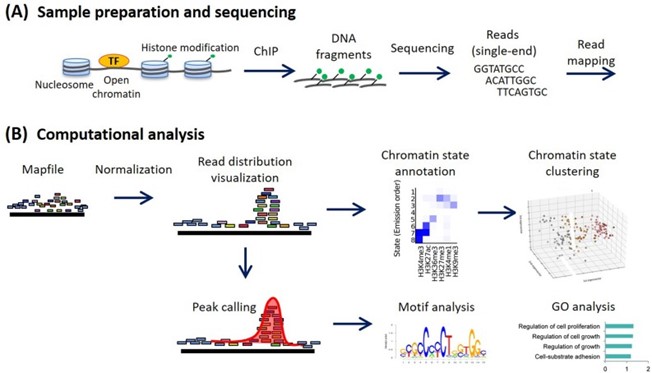 ChIP-seq analysis workflow. (Nakato, et al., 2021)
ChIP-seq analysis workflow. (Nakato, et al., 2021)
1. Data Analysis Methodology:
- ChIP-seq utilizes next-generation sequencing (NGS) to read and map DNA fragments acquired through immunoprecipitation. This approach provides unparalleled precision in the high-resolution mapping of protein-DNA interactions across the genome.
- ChIP-on-chip, on the other hand, involves hybridizing the enriched DNA fragments onto a DNA microarray. The detection of binding sites is based on the signal intensities from specific microarray regions, offering lower resolution than ChIP-seq and limited to pre-defined genomic segments.
2. Genomic Coverage:
- ChIP-seq provides a complete genome-wide analysis, detecting protein binding sites across many regulatory elements, including promoters, enhancers, and silencers. Its broad coverage often results in the identification of new binding sites.
- ChIP-on-chip is limited to the genomic regions chosen during the array's design. Even with arrays covering the entire genome, its scope is narrower compared to ChIP-seq, often missing novel sites not included in the chip design.
3. Complexity of Data Analysis:
- ChIP-seq involves a complex and resource-intensive data analysis process, with steps including data preprocessing, sequence alignment, peak identification, and annotation. The substantial data output requires advanced computational tools for thorough analysis.
- ChIP-on-chip analysis is more straightforward, focusing on evaluating and comparing probe signal intensities to pinpoint target binding regions, making the process less computationally demanding.
4. Flexibility and Application Range:
- ChIP-seq is exceptionally adaptable and suitable for an extensive range of research inquiries. Its strength lies in probing both known and unexplored binding sites, making it optimal for in-depth analysis of regulatory networks across the genome.
- ChIP-on-chip is particularly effective for investigating specific, predetermined genome areas, such as transcription factor binding sites or regulatory regions of selected genes, and is frequently used in detailed studies of these components.
5. Cost and Equipment Requirements:
- ChIP-seq generally incurs higher costs, necessitating access to high-throughput sequencing technologies and advanced data analysis platforms. Despite the decreasing cost of sequencing, it remains a pricier alternative compared to ChIP-on-chip.
- ChIP-on-chip is a more cost-effective approach, requiring only microarray technologies and basic hybridization equipment, making it a budget-conscious choice with lower technical requirements.
Conclusion:
- ChIP-seq: This high-throughput sequencing approach is well-suited for expansive genome-wide studies, offering superior resolution and the potential to identify novel protein-DNA binding interactions.
- ChIP-on-ChIP: This microarray-based strategy is tailored for the analysis of known regions, offering cost savings though with limitations concerning resolution and genomic coverage compared to ChIP-seq.
Service you may intersted in
For more details, please refer to the following articles:
Selecting the Appropriate ChIP-related Technology
When conducting ChIP experiments, researchers must choose among various ChIP technology variants tailored to specific experimental objectives. Each ChIP method offers distinct advantages and limitations, making them suitable for different research contexts. To elucidate the differences and guide the selection of the most appropriate method, the following comparison outlines the principal characteristics and distinctions of common ChIP technologies, including ChIP-chip, ChIP-seq, ChIP-PCR, and ChIP-qPCR.
| Technology Type |
ChIP-chip |
ChIP-seq (ChIP + NGS) |
ChIP-PCR |
ChIP-qPCR Quantitative Analysis |
| Principle |
Utilizes DNA microarray chips to analyze ChIP-enriched DNA fragments |
Employs high-throughput sequencing to analyze ChIP-enriched DNA fragments |
Amplifies enriched DNA fragments via PCR |
Quantitatively analyzes enriched DNA fragments via qPCR |
| Genomic Coverage |
Specific pre-determined genome regions |
Genome-wide |
Specific known gene regions |
Specific known gene regions |
| Resolution |
Lower, reliant on probe coverage on the chip |
Extremely high, allowing precise binding site localization |
Lower, constrained by PCR primer design |
Lower, constrained by PCR primer design |
| Data Analysis Complexity |
Relatively simple, based on signal intensity and matching results |
Highly complex, requiring specialized data analysis platforms |
Simple, using conventional PCR techniques |
Simple, using conventional qPCR techniques |
| Cost |
Relatively low, suitable for large-scale screening |
Higher, dependent on high-throughput sequencing platforms |
Low, suitable for small-scale experiments |
Low, suitable for small-scale experiments |
| Flexibility |
Limited to designed pre-determined regions on chips |
Highly flexible, applicable for genome-wide analysis |
Flexible, allowing selection of any known region |
Flexible, allowing selection of any known region |
| Application |
Analyzes protein binding within specific genomic regions |
Suitable for genome-wide protein-DNA binding site analysis |
Validates known protein-DNA binding sites |
Validates binding stronghold in specific gene regions |
| Advantages |
Cost-effective, ideal for studying known regions |
High resolution, capable of discovering novel binding sites |
Simple operation, suitable for small-scale studies |
Rapid, quantitative, ideal for validation studies |
| Limitations |
Limited coverage, unable to identify novel binding sites |
High cost, complex data analysis |
Limited to few known target regions |
Limited to few known target regions |
Conclusions:
- ChIP-chip: Best suited for large-scale analysis of protein-DNA interactions in known genomic regions. While data analysis is straightforward, its resolution and flexibility are limited.
- ChIP-seq: Offers comprehensive genome-wide protein-DNA interaction analysis with high resolution and flexibility, although at higher costs and complexity in data analysis.
- ChIP-PCR: Provides a simple, cost-effective approach for validating protein-DNA interactions in a limited number of known gene regions. It is highly flexible but offers low resolution.
- ChIP-qPCR Quantitative Analysis: Ideal for quantitative assessment of protein-DNA binding strength in specific gene regions. It is easy to operate and cost-effective but confined to known targets.
Through these considerations, researchers can align their choice of ChIP-related technology with their experimental aims, available resources, and the specificity required for their studies.
Conclusion
ChIP-on-chip, which integrates ChIP with genomic microarray technology, is a valuable methodology for investigating protein-DNA interactions and elucidating their functions in the regulation of gene expression. This technique allows for the enrichment and microarray-based analysis of protein binding sites, enabling high-throughput genomic investigations. It is especially effective in the examination of transcription factors, histone modifications, and a variety of other epigenetic markers. While ChIP-on-chip presents advantages in cost and equipment simplicity compared to ChIP-seq, it is characterized by comparatively lower resolution and flexibility, restricting its applicability to predefined genomic regions.
Overall, ChIP-on-chip represents an essential tool in the fields of epigenetics, gene regulation, and the study of disease mechanisms. However, researchers are advised to align their choice of ChIP-related methodologies with specific experimental objectives, financial considerations, and their capacity for data analysis to determine the most appropriate technology for their investigative needs.
References:
- Sandmann, T., Jakobsen, J. & Furlong, E. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1, 2839–2855 (2006). https://doi.org/10.1038/nprot.2006.383
- Nakato, Ryuichiro, and Toyonori Sakata. "Methods for ChIP-seq analysis: A practical workflow and advanced applications." Methods 187 (2021): 44-53. https://doi.org/10.1016/j.ymeth.2020.03.005
- Collas, P. The Current State of Chromatin Immunoprecipitation. Mol Biotechnol 45, 87–100 (2010). https://doi.org/10.1007/s12033-009-9239-8
- Makoukji, Joelle. "Chromatin immunoprecipitation assay for analyzing transcription factor activity at the level of peripheral myelin gene promoters." Psychiatric Disorders: Methods and Protocols (2019): 647-658. https://doi.org/10.1007/978-1-4939-9554-7_37
- Sen, Ilke, Alan Kavšek, and Christian G. Riedel. "Chromatin Immunoprecipitation and Sequencing (ChIP‐seq) Optimized for Application in Caenorhabditis elegans." Current Protocols 1.7 (2021): e187. https://doi.org/10.1002/cpz1.187


 Sample Submission Guidelines
Sample Submission Guidelines
 Schematical overview of ChIP-on-chip experiments. (Sandmann, T., et al., 2006)
Schematical overview of ChIP-on-chip experiments. (Sandmann, T., et al., 2006) The chromatin immunoprecipitation (ChIP) assay and various methods of analysis. (Collas, et al., 2010)
The chromatin immunoprecipitation (ChIP) assay and various methods of analysis. (Collas, et al., 2010) ChIP-seq analysis workflow. (Nakato, et al., 2021)
ChIP-seq analysis workflow. (Nakato, et al., 2021)