The exploration of complex phenotypic traits and their genetic underpinnings has been revolutionized through Genome-Wide Association Studies (GWAS). Since their initial application to human disorders in 2005, these analytical methods have generated significant insights across multiple scientific disciplines. Our examination focuses on three key domains where GWAS has demonstrated particular utility: agricultural advancement, conservation of biological diversity, and investigations into human genetic variation. We present relevant examples to demonstrate the practical implementation of this genomic approach.
Applications of GWAS in Agriculture
GWAS serves as a powerful tool in agriculture by facilitating gene discovery for crop improvement, enhancing our understanding of genetic diversity for conservation efforts, and optimizing animal breeding practices for better productivity and health outcomes.
Gene Discovery and Breeding Improvement
GWAS have revolutionized the identification of key genes that significantly influence crop yield, quality, and resistance to various stresses. For example, in maize, GWAS has pinpointed genetic loci associated with traits such as kernel number and drought tolerance. A notable study identified SNPs linked to oleic acid content in maize kernels, which can enhance the nutritional quality of the crop while also improving its resistance to environmental stressors(Sahito,et.al,2024).
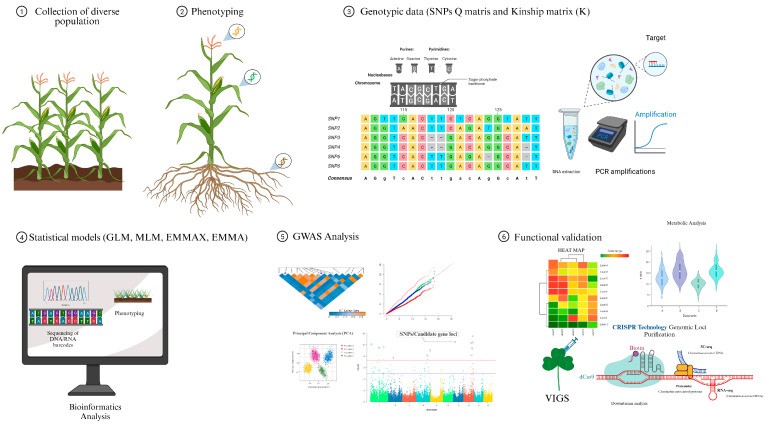 Fig. 1. Basic GWAS approach in maize.(Sahito, et.al, 2024)
Fig. 1. Basic GWAS approach in maize.(Sahito, et.al, 2024)
Crop Genetic Diversity Research
Understanding the genetic foundation of crops is crucial for promoting variety conservation and improving resource management. For example, a GWAS performed on different sorghum lines identified notable genetic diversity associated with traits like plant height and flowering time. This research contributes to conservation strategies while advancing breeding methods by pinpointing genetic markers that can enhance crop resilience and adaptability (Kasule et al., 2024). Furthermore, investigations on Arabidopsis have demonstrated how GWAS can reveal the genetic pathways responsible for flowering time adjustments to environmental factors, a key aspect of developing crops that can flourish in diverse climate conditions (Nakano et al., 2020).
Animal Breeding
GWAS has also made substantial contributions to animal breeding by enhancing production efficiency and improving the quality of animal products. For example, a study on livestock identified genomic regions associated with traits like milk production and disease resistance in dairy cattle. By leveraging GWAS findings, breeders can select animals with desirable genetic profiles, thereby increasing overall herd productivity and health(Pavan,et.al,2020).
You may interested in
Resource
Applications of GWAS in Biodiversity Research
GWAS serves as a powerful tool in biodiversity research by enhancing our understanding of species evolution and adaptation while providing critical insights for conservation efforts aimed at preserving genetic diversity and species resilience.
Species Evolution and Adaptation Studies
GWAS have played a crucial role in revealing the genetic mechanisms that underlie species' adaptation to their environments. For example, research on Arabidopsis thaliana highlighted how certain alleles became more common in populations as a result of adaptive responses to local climate factors. The study identified 821 single nucleotide polymorphisms (SNPs) that were strongly linked to various environmental variables, demonstrating that adaptations typically result from gradual changes in allele frequencies rather than from large, impactful mutations. This observation underscores the ability of GWAS to shed light on the intricate genetic foundation of adaptation, providing a more comprehensive understanding of evolutionary processes across different species (De La Torre et al., 2019).
Conservation Genetics
In the realm of conservation genetics, GWAS are essential for evaluating the genetic diversity of threatened species and developing effective conservation measures. A notable example is a GWAS performed on the critically endangered Florida panther, which uncovered low levels of genetic diversity and pinpointed specific loci linked to traits important for survival and reproduction (Saremi et al., 2019). This valuable information has significantly influenced management strategies, including breeding initiatives designed to enhance genetic diversity by incorporating individuals from different populations.
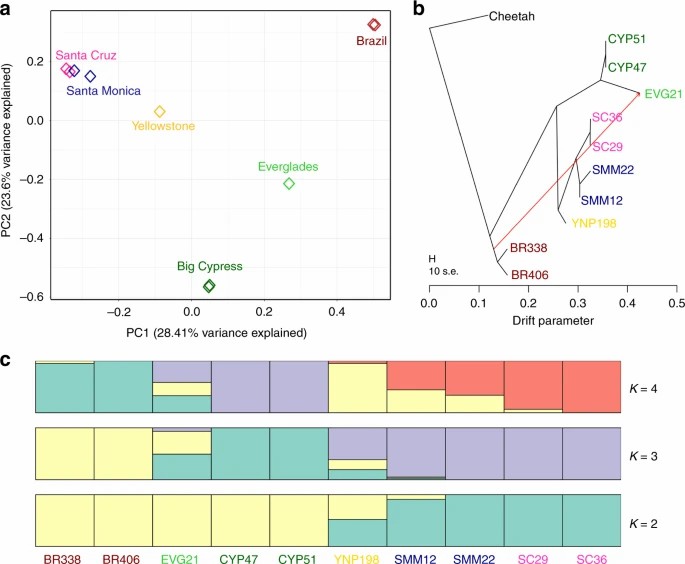 Fig. 2. Stratification of pumas based on the geographic population.(Saremi,et.al,2019)
Fig. 2. Stratification of pumas based on the geographic population.(Saremi,et.al,2019)
Applications of GWAS in Human Population Genetics Research
GWAS plays an essential role in human population genetics by clarifying the structure and migration patterns of populations. It also offers valuable insights into the genetic determinants that affect health and disease in various populations.
Population Structure Analysis
GWAS has significantly advanced our understanding of human population structure, revealing insights into the history of human origins, migrations, and admixture. For example, the Geographic Population Structure (GPS) algorithm utilizes genetic data to accurately predict the geographical origins of individuals. In a study involving over 200 Sardinian villagers, GPS successfully placed 83% of individuals in their country of origin, with many accurately located within 50 km of their villages(Elhaik,et.al,2014).Additionally, research on the genetic diversity of Native American populations has shown distinct genetic signatures that reflect their unique migratory paths and adaptations to diverse environments across the Americas, further demonstrating the power of GWAS in tracing human history(Gibson G,2018).
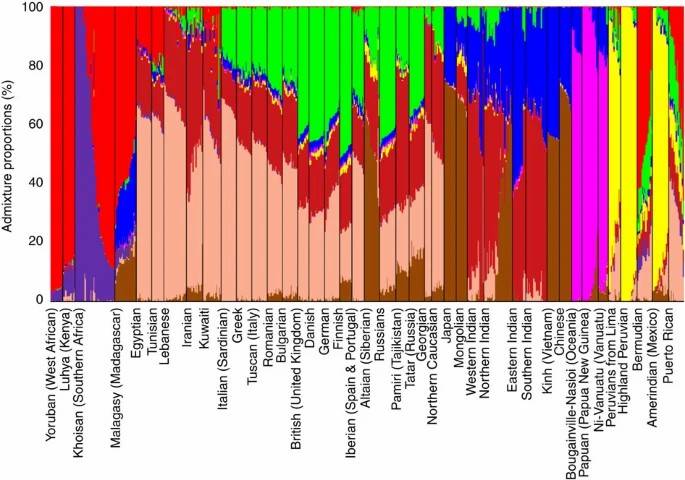 Fig. 3. Admixture analysis of worldwide populations and subpopulations.(Elhaik,et.al,2014)
Fig. 3. Admixture analysis of worldwide populations and subpopulations.(Elhaik,et.al,2014)
Genetic Epidemiology Studies
Through extensive research initiatives, GWAS have significantly advanced our understanding of how genes impact disease patterns and overall health outcomes across populations. The identification of genetic risk factors for type 2 diabetes (T2D) represents a significant achievement in this field. Researchers conducting large-scale genomic analyses successfully pinpointed multiple single nucleotide polymorphisms (SNPs) that correlate with T2D susceptibility. These investigations demonstrated varying frequencies of risk-associated alleles among different ethnic groups, which aligns with documented geographical patterns of T2D prevalence. Notably, comparative analyses revealed that specific genetic variants linked to increased T2D risk occur more frequently in Europeans compared to populations of African descent, suggesting an interplay between ancestral background and environmental conditions in determining disease susceptibility (Simons et al., 2018).
Applications of GWAS in Drug Development
The applications of GWAS in drug development include identifying novel therapeutic targets by uncovering associations between genetic variants and diseases, as well as elucidating the genetic basis of adverse drug reactions, thereby enhancing drug safety and therapeutic potential.
Drug Target Discovery
GWAS play a pivotal role in identifying novel therapeutic targets by correlating genetic variants with diseases. For instance, the interleukin-6 receptor (IL-6R) has been established as a target for tocilizumab, a drug used primarily for rheumatoid arthritis. GWAS findings have also linked IL-6R to coronary heart disease, suggesting potential for expanding its use to treat cardiovascular conditions as well(Gordillo, et.al, 2024).Another example is the interleukin-23 receptor (IL-23R), initially targeted for psoriasis, which was later investigated and approved for Crohn's disease after GWAS identified relevant genetic signals(Simon ,et.al, 2016).
Drug Safety Assessment
GWAS are instrumental in understanding the genetic underpinnings of adverse drug reactions (ADRs). For example, research has identified specific human leukocyte antigen (HLA) genotypes that predict susceptibility to drug-induced liver injury from medications like abacavir and carbamazepine. In the case of carbamazepine, the HLA-A*3101 allele was found to significantly increase the risk of severe skin reactions, prompting more cautious prescribing practices for patients with this genetic marker(Ozeki, T,et.al, 2011).
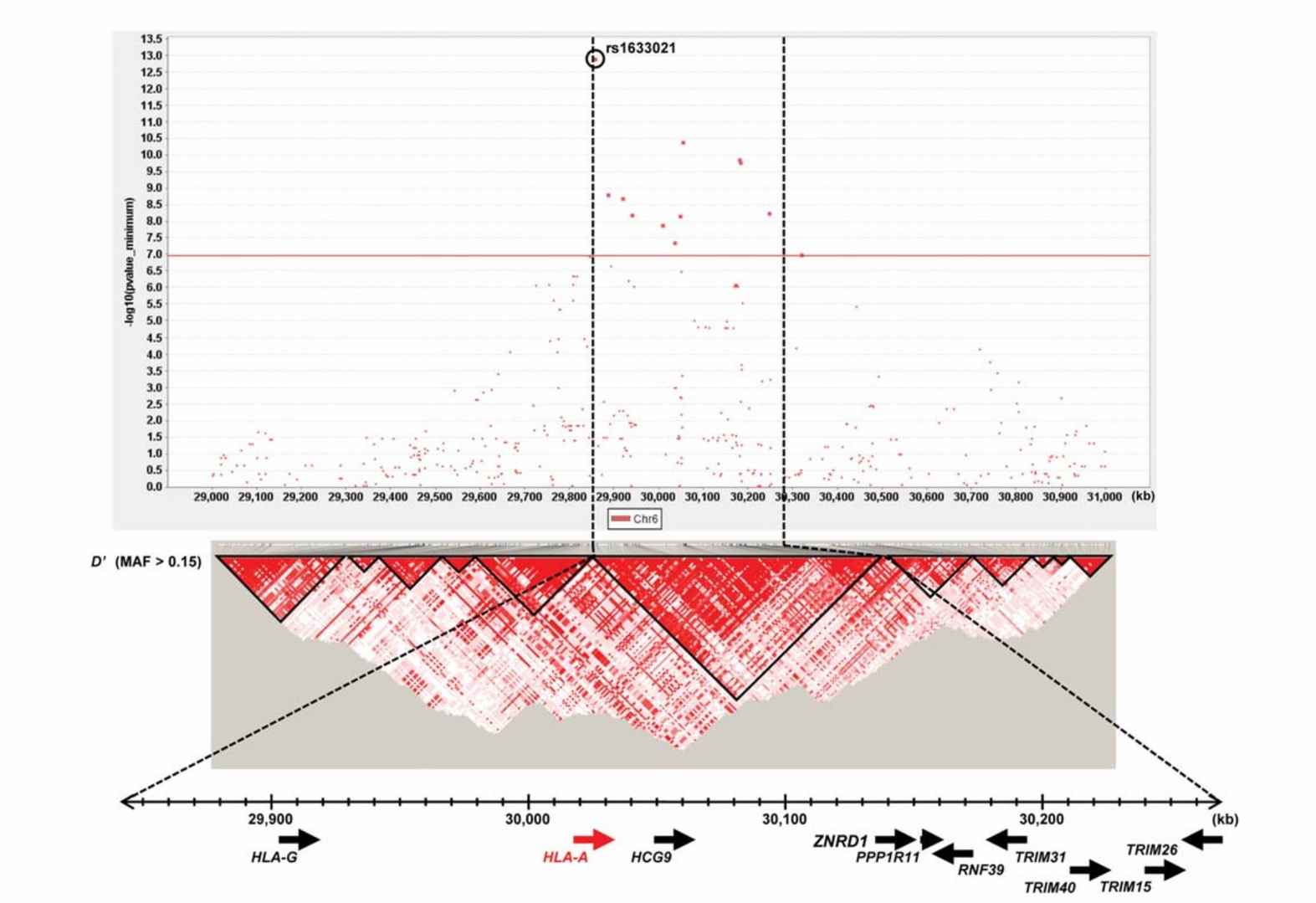 Fig. 4. LD map and genomic structure of an HLA-A region.(Ozeki, T,et.al, 2011)
Fig. 4. LD map and genomic structure of an HLA-A region.(Ozeki, T,et.al, 2011)
Applications of GWAS in Biological Big Data Mining
GWAS applications in biological big data mining are significantly enhanced through the integration of multi-omics data and the development of innovative bioinformatics methods, leading to deeper insights into genetic mechanisms and improved research outcomes.
Data Integration and Analysis Techniques
The integration of multi-omics data is crucial for enhancing the depth and breadth of research in biological big data mining. For example, a study analyzing metabolic traits in Brassica napus (canola) utilized multi-omics approaches, combining genomics, transcriptomics, and metabolomics to identify candidate genes associated with growth variation. By integrating high-throughput phenotyping data with RNA sequencing and metabolite profiling, researchers discovered over 61,000 quantitative trait loci (QTL), significantly improving the understanding of the genetic basis of complex traits in canola. This comprehensive approach not only highlighted key genes involved in early biomass accumulation but also demonstrated how multi-omics integration can reveal intricate relationships between different biological layers, ultimately leading to more effective breeding strategies for crop improvement(Knoch, et.al, 2024).
Bioinformatics Method Innovation
The development of new algorithms and tools is essential to address challenges in GWAS data analysis. For instance, machine learning techniques have been increasingly applied to enhance GWAS results by improving statistical power and identifying epistatic interactions among genetic variants.
Conclusion
GWAS serve as a robust instrument in numerous domains, such as agriculture, biodiversity, and human population genetics. This methodology facilitates the identification of genes that can enhance crop quality, supports research on biodiversity, and refines practices in animal breeding. In the context of human genetics, GWAS aids in comprehending population structures and the genetic factors associated with diseases. The incorporation of multi-omics data along with advancements in bioinformatics has broadened the scope of GWAS applications, yielding more profound insights into complex traits. In summary, GWAS propels advancements in scientific inquiry and practical implementations across various fields.
References:
- Sahito, J. H., Zhang, H., et.al. (2024). Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize. International journal of molecular sciences, 25(3), 1918. https://doi.org/10.3390/ijms25031918
- Kasule, F., Alladassi, B. M. E., et.al. (2024). Genetic diversity, population structure, and a genome-wide association study of sorghum lines assembled for breeding in Uganda. Frontiers in plant science, 15, 1458179. https://doi.org/10.3389/fpls.2024.1458179
- Nakano, Y., & Kobayashi, Y. (2020). Genome-wide association studies of agronomic traits consisting of field-and molecular-based phenotypes. Reviews in Agricultural Science, 8, 28-45. https://doi.org/10.7831/ras.8.0_28
- Pavan, S., Delvento, C., et.al. (2020). Recommendations for Choosing the Genotyping Method and Best Practices for Quality Control in Crop Genome-Wide Association Studies. Frontiers in genetics, 11, 447. https://doi.org/10.3389/fgene.2020.00447
- De La Torre, A. R., Wilhite, B., & Neale, D. B. (2019). Environmental Genome-Wide Association Reveals Climate Adaptation Is Shaped by Subtle to Moderate Allele Frequency Shifts in Loblolly Pine. Genome biology and evolution, 11(10), 2976–2989. https://doi.org/10.1093/gbe/evz220
- Saremi, N.F., Supple, M.A., Byrne, A. et al. Puma genomes from North and South America provide insights into the genomic consequences of inbreeding. Nat Commun 10, 4769 (2019). https://doi.org/10.1038/s41467-019-12741-1
- Elhaik, E., Tatarinova, T., Chebotarev, D. et al. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun 5, 3513 (2014). https://doi.org/10.1038/ncomms4513
- Gibson G. (2018). Population genetics and GWAS: A primer. PLoS biology, 16(3), e2005485. https://doi.org/10.1371/journal.pbio.2005485
- Simons, Y. B., Bullaughey, K., Hudson, R. R., & Sella, G. (2018). A population genetic interpretation of GWAS findings for human quantitative traits. PLoS biology, 16(3), e2002985. https://doi.org/10.1371/journal.pbio.2002985
- Gordillo, M., Schmidt, A.F., Warwick, A. et al. Disease coverage of human genome-wide association studies and pharmaceutical research and development. Commun Med 4, 195 (2024). https://doi.org/10.1038/s43856-024-00625-5
- Simon, E. G., Ghosh, S., Iacucci, M., & Moran, G. W. (2016). Ustekinumab for the treatment of Crohn's disease: can it find its niche?. Therapeutic advances in gastroenterology, 9(1), 26–36. https://doi.org/10.1177/1756283X15618130
- Ozeki, T., Mushiroda, T., et.al. (2011). Genome-wide association study identifies HLA-A*3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Human molecular genetics, 20(5), 1034–1041. https://doi.org/10.1093/hmg/ddq537
- Knoch, D., Meyer, R. C., et.al. (2024). Integrated multi-omics analyses and genome-wide association studies reveal prime candidate genes of metabolic and vegetative growth variation in canola. The Plant journal : for cell and molecular biology, 117(3), 713–728. https://doi.org/10.1111/tpj.16524


 Sample Submission Guidelines
Sample Submission Guidelines
 Fig. 1. Basic GWAS approach in maize.(Sahito, et.al, 2024)
Fig. 1. Basic GWAS approach in maize.(Sahito, et.al, 2024) Fig. 2. Stratification of pumas based on the geographic population.(Saremi,et.al,2019)
Fig. 2. Stratification of pumas based on the geographic population.(Saremi,et.al,2019) Fig. 3. Admixture analysis of worldwide populations and subpopulations.(Elhaik,et.al,2014)
Fig. 3. Admixture analysis of worldwide populations and subpopulations.(Elhaik,et.al,2014) Fig. 4. LD map and genomic structure of an HLA-A region.(Ozeki, T,et.al, 2011)
Fig. 4. LD map and genomic structure of an HLA-A region.(Ozeki, T,et.al, 2011)