We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Navigation
- Home
- Sequencing
- Genomics Sequencing
- Complete Plasmid/Phage Sequencing
- Mitochondrial DNA (mtDNA) Sequencing
- Viral Genome Sequencing
- Whole Genome Sequencing
- Whole Exome Sequencing
- Targeted Region Sequencing
- Amplicon Sequencing Services
- Chloroplast DNA (cpDNA) Sequencing
- TCR-Seq
- Shallow Whole Genome Sequencing
- Human Mitochondrial DNA (mtDNA) Sequencing
- Long-Read Sequencing
- ctDNA Sequencing Services
- Gene Panel Sequencing Service
- Long Amplicon Analysis (LAA)
- Animal/Plant Exome Sequencing Service
- Transcriptomics
- Bacterial RNA Sequencing
- RNA-Seq
- Small RNA Sequencing
- LncRNA Sequencing
- CircRNA Sequencing
- Ribosome Profiling (Ribo-seq)
- Total RNA Sequencing
- Targeted RNA Sequencing
- Degradome Sequencing
- Exosomal RNA Sequencing
- Ultra Low RNA Sequencing
- Dual RNA-seq
- microRNA Sequencing Service
- mRNA Sequencing Service
- Epigenomics
- Targeted Bisulfite Sequencing
- EM-seq Service
- Reduced Representation Bisulfite Sequencing
- Whole Genome Bisulfite Sequencing (WGBS)
- MeDIP Sequencing (MeDIP-Seq)
- ChIP-Seq
- MeRIP Sequencing (m6A Analysis)
- RIP-Seq
- ATAC-Seq
- NGS-BSP
- DNA 6mA Sequencing
- DAP-Seq Service (DNA affinity purification sequencing)
- 5mC/5hmC Sequencing
- oxBS-seq
- hMeDIP-seq
- Nanopore RNA Methylation Sequencing Service
- RNA Methylation Sequencing Service
- 2'-O-RNA Methylation Sequencing Service
- PacBio SMRT Sequencing
- Microbiome
- Single-Cell Sequencing
- Nanopore Sequencing
- Other Services
- CRISPR Sequencing
- CRISPR Screen Sequencing
- CRISPR Off-Target Validation
- Antibody Screening Sequencing (Phage Display Library Screening)
- Immune Repertoire Sequencing
- Sanger Sequencing
- Pre-made Library Sequencing
- Lentiviral/Retroviral Integration Sites Analysis
- AAV Genome Sequencing
- AAV (Adeno-Associated Virus) Integration Site Analysis
- 10x Spatial Transcriptome Sequencing Service
- HLA Typing
- Genomics Sequencing
- Genotyping
- Population Genetics
- Bioinformatics
- Microarray
- Applications
- Company
- Get Your Instant Quote


 Sample Submission Guidelines
Sample Submission Guidelines
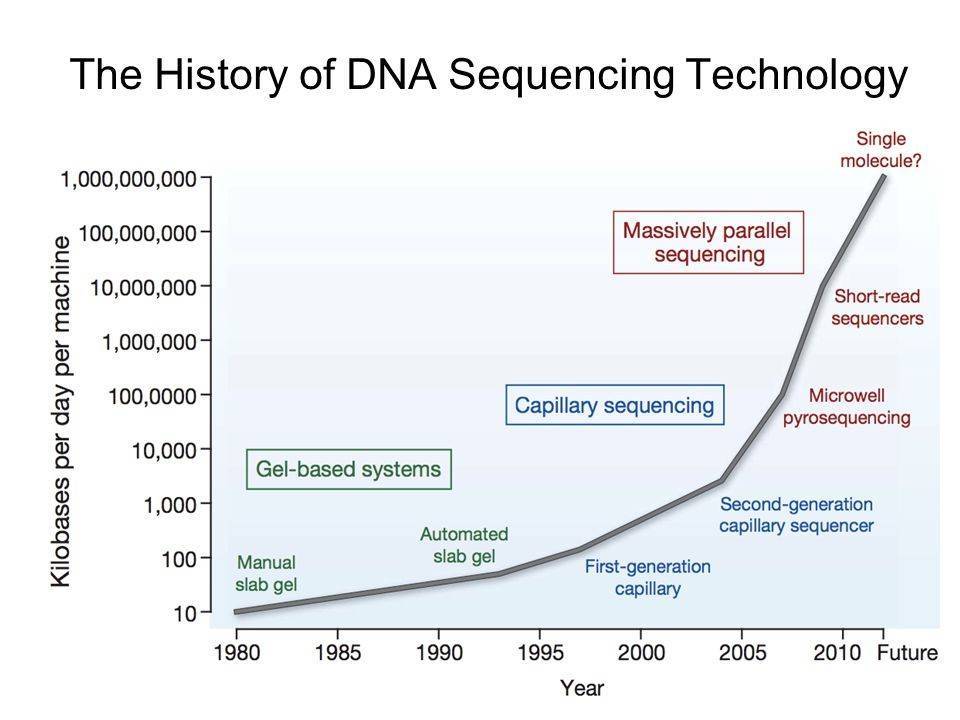 Figure 1. The history of DNA sequencing technology.
Figure 1. The history of DNA sequencing technology.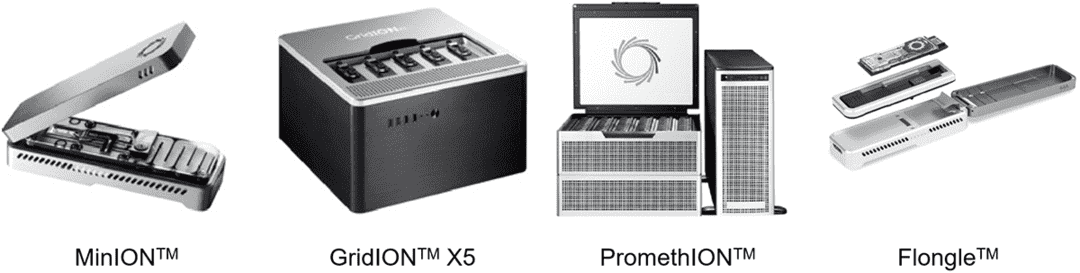 Figure 2. Nanopore sequencing platforms at ONT.
Figure 2. Nanopore sequencing platforms at ONT.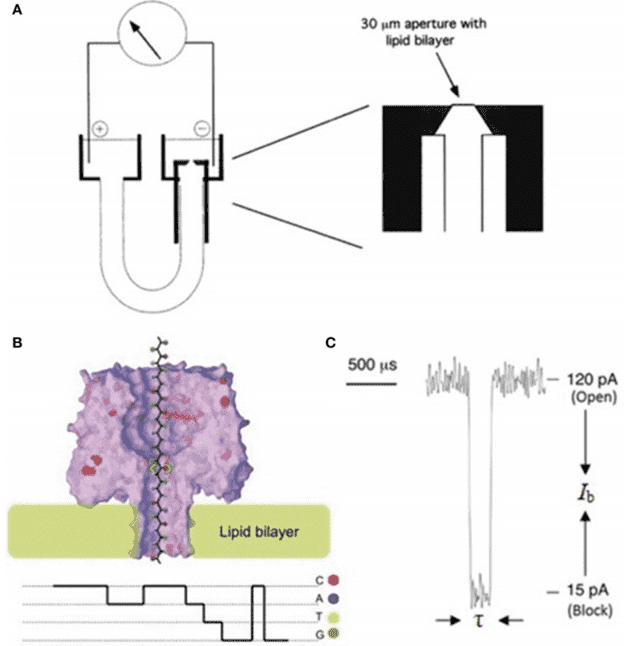 Figure 3. Schematic illustration of a nanopore sequencing device.
Figure 3. Schematic illustration of a nanopore sequencing device.