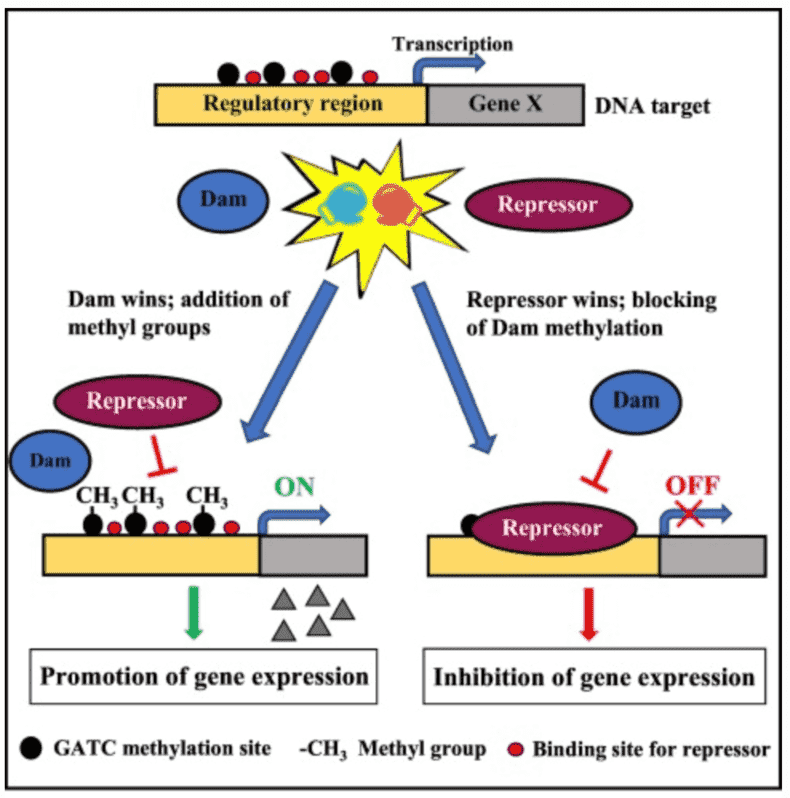
Out of the multiple defense mechanisms of bacteria against phages, the most broadly studied are the methylation-based R-M systems. Restriction-modification (R-M) systems confer resistance to phage infection through a methyltransferase (MTase) which methylates a specific sequence of self-DNA and a cognate restriction endonuclease that discriminates and then destroys foreign invasive DNA. To distinguish self-DNA from foreign DNA, bacteria have developed diverse R-M systems to perform DNA methylation and generate N4-methyl-cytosine, N6-methyl-adenine, and C5-methyl-cytosine in the process.
Phosphorothioation is a post-replicative modification on the DNA backbone and is widespread and quantized in bacterial genomes. Phosphorothioate (PT) modification happens when the non-bridging oxygen atom in the phosphate group of the DNA sugar-phosphate backbone is replaced with sulfur. It was initially utilized as an artificially protect oligonucleotides against nuclease degradation. With further studies, it was recently discovered in bacteria that PT modification occurs naturally and it surpasses our understanding of R-M. The PT system is believed to be more complicated than DNA methylation because it involves sequence-selective and stereospecific incorporation of sulfur through the action of five Dnd proteins. Initiation of PT modification processes depends on the presence of the dnd gene clusters. The PT modification system is comprised of two mutually exclusive parts: the modification (M) component which is the gene product of the dndABCDE cluster function conferring the PT modification and the downstream restriction (R) component which is the dndFGH products recognizing and hydrolyzing non-PT-protected foreign DNA. Turning-off of PT modifications showed an increase in double-stranded DNA breaks and oxidative stress in Salmonella and Pseudomonas, respectively). The table below shows the cluster groups and their functions.
Cluster group members of modification and restriction components of the PT system and their functions (Tong, 2018)
| Cluster Member |
Functions |
| dndA |
Performs cysteine desulfurase activity |
| dndB |
Binds to the promoter region of the dnd operon to regulate the dnd gene transcription |
| dndC |
Possesses ATP pyrophosphatase activity |
| dndD |
Shows ATPase activity and provides energy during the incorporation of sulfur |
| dndE |
Facilitates preferential binding to nicked double-strand DNA in vitro |
| dndF |
Restriction function |
| dndG |
| dndH |
Sequencing Technologies for Studying Phosphorothioation
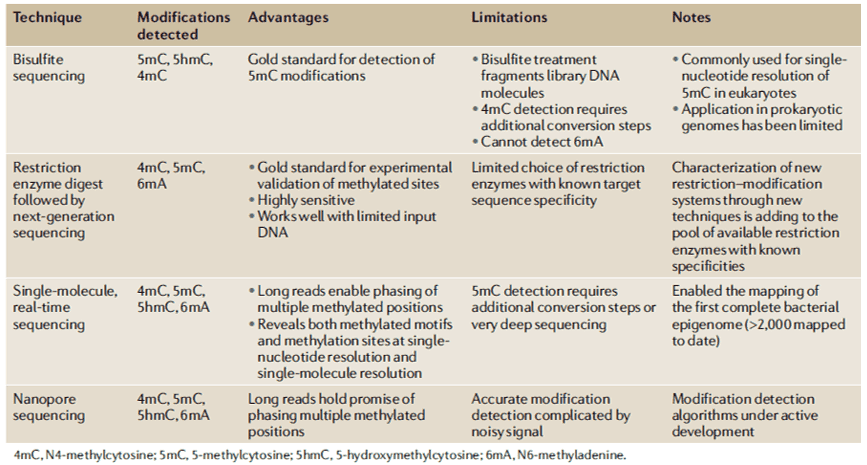
Our understanding of the biological functions of PT modifications remains limited, and these limitations include sequence context, genomic frequency, and relationship to dnd gene cluster diversity. Metagenomic analysis of PT modifications reveals a unique ecological distribution, and phylogenetic comparisons of dnd genes and PT sequence context strongly support horizontal transfer of dnd genes. Most studies adopt the single-molecule, real-time (SMRT) sequencing technology to map PT sites across bacterial genomes. Furthermore, RNA-seq analysis was conducted to characterize cellular responses and to determine whether PT modifications play a role in the epigenetic control of the expression of certain genes. The liquid chromatography with tandem mass spectrometry (LC-MS/MS) method is being used to define the details in the PT sequence and quantify PT modification frequencies in the bacterial genome known to harbor dnd gene clusters.
References
- Tong T, Chen S, Wang L, et al. Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proceedings of the National Academy of Sciences. 2018, 115(13).
- Wang L, Jiang S, Deng Z, et al. DNA phosphorothioate modification—a new multi-functional epigenetic system in bacteria. FEMS microbiology reviews. 2019, 43(2).
- Chen C, Wang L, Chen S, et al. Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes. Proceedings of the National Academy of Sciences. 2017, 114(17).
- Wang L, Chen S, Vergin KL, et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proceedings of the National Academy of Sciences. 2011, 108(7).
Related Sections
Services: