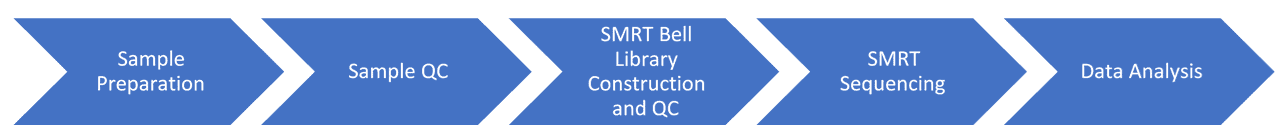
Introduction to Our Microbial DNA Phosphorothioation Mapping Platform

Single Molecular Real-Time (SMRT) sequencing uses an advanced flow cell with many hundreds of transparent bottoms of individual picolitre wells, zero-mode waveguides (ZMW). The polymerase is placed at the bottom of the well and enables the progression of the DNA strand through the ZMW. As a result, the structure can concentrate on a single molecular element. SMRT sequencing enables fluorescently tagged nucleotides that are modulated along with individual DNA layout molecules to be imaged in real-time. When the template and polymerase detach, the sequencing reaction terminates. The PacBio instrument's average read length is estimated at 2 kb, and some reads may exceed 20 kb. For de novo arrays of novel genomes that can stretch many more repeats and bases, longer reads are particularly useful.
Highly repetitive elements discovered in both eukaryotic and prokaryotic genomes present a challenge to the assembly of genomes and make it difficult to thoroughly analyze repetitive sequences. Long-read sequencing provides reads exceeding several or tens of kilobases (kbs) that can spread complicated or repetitive regions with a single continuous reading, enabling these large structural features to be resolved. It can also show where methylated bases take place, in addition to significantly longer and extremely reliable DNA sequences from individual unamplified molecules, offering functional data about genome-encoded DNA methyltransferases. In research of de novo genomics, metagenomics, transcriptomics, and epigenetics, PacBio SMRT sequencing has distinct benefits.