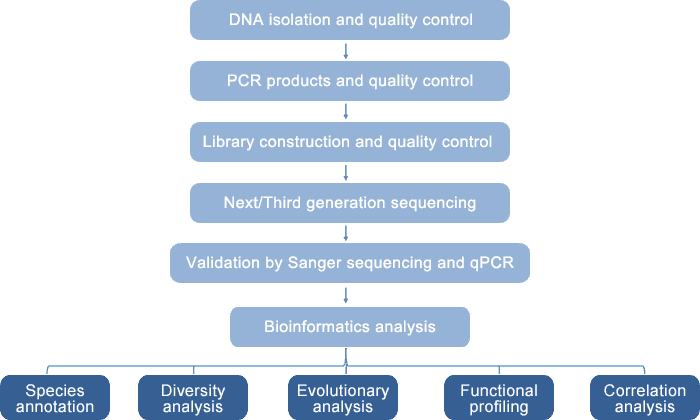
Our integrated and novel platform enables high-throughput and reliable 18S/ITS analysis for eukaryotic diversity detection with a combination of sequencing technology (Sanger, Illumina MiSeq/HiSeq, and PacBio SMRT sequencing) and quantitative PCR (q-PCR). The principle of this platform is based on the role of 18S rRNA and ITS as molecular markers in eukaryotes. This platform allows both short-read 18S/ITS sequencing by Illumina MiSeq/HiSeq (such as the amplification and sequencing of V4 region of 18S rRNA) and full-length 18S/ITS sequencing by PacBio SMRT technology. The data need to be further validated by Sanger sequencing and q-PCR with genus- and species-specific primers. Quantitative PCR can also be used to quantify microbial strains. Based on our eukaryotic diversity analysis platform, we provide provide eukaryotic 18S rRNA sequencing and fungal ITS sequencing.
Our platform has supported a variety of fields, including basic research, agriculture, environment, as well as pharmaceutical and food & beverage industries, etc. Our featured eukaryotic diversity analysis services include soil actinomycetes diversity analysis and clinical microbiology. In addition to eukaryotic diversity analysis platform, prokaryotic diversity analysis platform is also available for comprehensive microbial diversity detection by using 16S rRNA as molecular marker.