CD genomics can help you analyze genetic variation efficiently to meet diverse research needs. By utilizing Affymetrix and Illumina array platforms, we support high-throughput, multiplex processing to meet diverse research needs., delivering high-quality data at a low per-sample cost.
What is a SNP Microarray Test
Single Nucleotide Polymorphism (SNP) microarray represents a sophisticated, high-throughput genomic technology adept at detecting and analyzing single nucleotide polymorphisms across the human genome. SNPs constitute the most prevalent form of genetic variation among individuals, characterized by the alteration of a single nucleotide within the DNA sequence. These tiny genomic alterations can exert significant influence on a wide spectrum of biological traits, encompassing disease susceptibility and pharmacogenomic response.
SNP Microarray Detection Principles:
- Chip Composition: The microarray primarily consists of fiber optic strands and nanomaterials, such as silicon beads, which form the core components of the chip.
- Technical Principles: The chip integrates numerous known sequence probes. These probes hybridize with fluorescently labeled DNA molecules from samples to detect hybridization signals. This process facilitates the acquisition of sequence information.
CD Genomics harnesses state-of-the-art SNP microarray technology to deliver precise and comprehensive genetic analyses. By employing high-density array platforms, CD Genomics provides detailed insights into genetic variations, which may elucidate the molecular underpinnings of diverse phenotypes and medical conditions. This advanced technology facilitates the identification of critical genomic loci responsible for disease predisposition and therapeutic responsiveness, potentially guiding preventative strategies and personalized medical interventions.
The Introduction to SNP Microarray Service
Equipped with advanced variant detection and SNP genotyping technologies, CD Genomics offers a suite of genotyping services that deliver high-quality data at competitive per-sample costs. SNP microarrays are an invaluable tool for researchers, facilitating the exploration of genetic variants—including SNPs and significant structural changes—across a spectrum of applications, from initial discovery to routine screening.
Both Affymetrix and Illumina genotyping microarrays are known for their reliable performance, making them suitable for precision medicine initiatives, clinical and translational research, pharmacology, consumer screening, and agricultural applications. Depending on the species and the specific SNP selection, a range of genotyping strategies can be employed to meet diverse research needs.
Table 1. Our available automated SNP genotyping platforms.
| Platform | Applications | |
|---|---|---|
 |
GeneTitan instrument together with Axiom array plates |
|
 |
GeneChip Scanner 3000 7G System together with Affymetrix GeneChip |
|
 |
Illumina iScan system together with Illumina BeadChips |
|
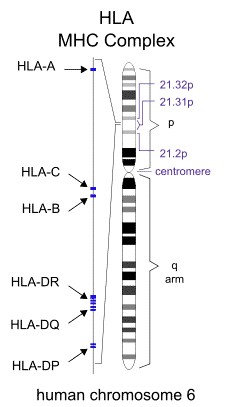 Figure 1. Diagram of the location of HLA-encoding genes.
Figure 1. Diagram of the location of HLA-encoding genes.
Advantages of Our SNP Microarray Service
- Custom, flexible, and scalable
- High call rates > 99%, and high accuracy
- Cost-effective and high-throughput
- High Resolution and Accuracy
- Broad Coverage and Flexibility
- Rapid Turnaround Time
- Comprehensive Data Interpretation
- Identifies SNPs in targeted or whole genome scale
- In addition to SNPs, other genetic differences, such as copy number variations, can be measured
- Applications in biomarker discovery and validation, clinical testing, GWAS, pharmacogenomics, forensics, and breed discrimination
Applications of SNP Microarray
- Genetic Research: SNP microarrays provide a detailed view of genetic variations linked to specific diseases and traits, driving forward our knowledge of genetic influences and mechanisms.
- Personalized Medicine: By analyzing individual genetic profiles, SNP microarrays enable customized treatment plans, improving drug effectiveness and reducing the risk of adverse drug reactions.
- Population Genetics and Evolutionary Studies: These arrays offer insights into genetic diversity and evolutionary changes across different populations, helping to trace migration patterns and evolutionary developments.
- Clinical Diagnostics: SNP microarrays are instrumental in diagnosing genetic conditions and evaluating disease risks, facilitating early intervention and tailored healthcare strategies.
- Agriculture and Animal Breeding: In agriculture, SNP microarrays are used to enhance desirable traits in crops and livestock, leading to better yields and improved resistance to diseases.
SNP Microarray Workflow
The general workflow for SNP Microarray is outlined below. We have three well-recognized genotyping platforms (Table 1) for genome-wide or targeted identification of SNPs and copy number variations in a high-throughput and affordable manner. Our highly experienced expert team executes quality management, following every procedure to ensure confident and unbiased results.
Sample Requirements
|
|
Click |
SNP Genotyping
|
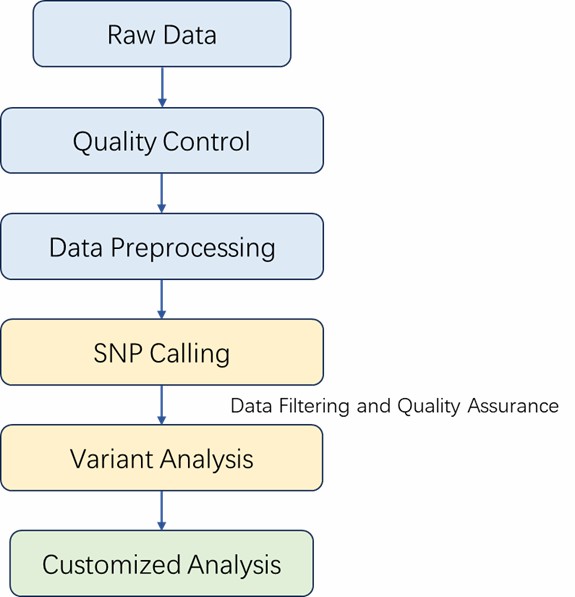
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in SNP Microarray for your writing (customization)
Our full-service offering encompasses the entire workflow, from quality control of genomic DNA to the precise determination of genotypes. Additionally, CD Genomics provides the capability to design custom microarrays tailored to meet unique project requirements. For further inquiries or additional specifications, please do not hesitate to contact our team.
Partial results are shown below:
1.The introduction of Illumina and Affymetrix genotyping platforms.
Table 1. Affymetrix and Illumina genotyping solutions.
| Affymetrix genotyping solutions | The Affymetrix SNP microarray applies a combination of photolithography and combinatorial chemistry to directly synthesize oligonucleotides on a glass surface. Affymetrix arrays can achieve very high density to accommodate millions of probes on a single chip. |
| Illumina genotyping solutions | Illumina's unique BeadArray technology is based on 3-micron silica beads that self-assemble in micro-wells on planar silica slides. Each bead is covered with hundreds of thousands of copies of a specific oligonucleotide, which acts as the capture sequence. Accuracy and reliability of the assays are usually above 99.5%. |
2.How to choose the most suitable genotyping platform for my specific needs?
The choice of genotyping strategies depends on the species, the purpose, the number of SNPs per sample, and the number of samples. For different cases, we recommend different genotyping technologies.
1. For genome-wide SNP genotyping, we recommend Illumina BeadChips, Affymetrix GeneChips or NGS.
2. For gene-wide SNP genotyping, such as fine mapping or haplotyping of candidate regions we recommend NGS or array-based technologies.
3. For the analysis of individual SNPs, we recommend Real-time PCR assays, Sanger Sequencing, Illumina BeadChips, Affymetrix technology, or NGS.
3.What arrays are available at CD Genomics?
At CD Genomics, we have both ready-made and custom arrays for your SNP genotyping projects.
Table 2. Our available arrays for SNP genotyping.
| Humans | Animals | Plants |
|
|
|
Identification of Quantitative Trait Loci for Clubroot Resistance in Brassica oleracea With the Use of Brassica SNP Microarray
Journal: Frontiers in Plant Science
Impact factor: 4.1
Published: 18 June 2018
Background
Clubroot, caused by Plasmodiophora brassicae, severely impacts cabbage by inducing root galls that hinder water and nutrient uptake, leading to abnormal growth. Control is challenging due to the pathogen's persistence in soil, making the development of resistant cultivars essential. Identifying clubroot resistance (CR) quantitative trait loci (QTLs) in Brassica species, including B. oleracea, is crucial for breeding resistant varieties. Recent research utilized a 60K Brassica SNP microarray to map CR QTLs in an F2 population derived from a resistant cabbage line, aiding in the development of clubroot-resistant cabbages.
Materials & Methods
Sample Preparation
- Cabbage
- DNA extraction
Method
- Genotyping
- Bead Chip Array (Infinium)
- Genetic map construction
- QTL analysis
Results
1. Phenotypic Performance of Parents and F2 Lines
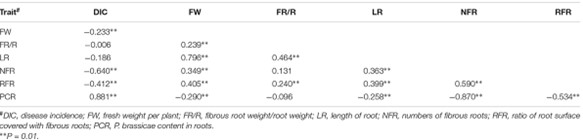
'GZ87' demonstrated complete resistance to clubroot, while '263' was highly susceptible. The F2 lines showed significant variability in traits, with strong correlations identified between disease incidence and certain traits like PCR and NFR.
TABLE 1. The phenotypic performance of clubroot resistance associated traits in the parental lines and the F2 population of Brassica oleracea.
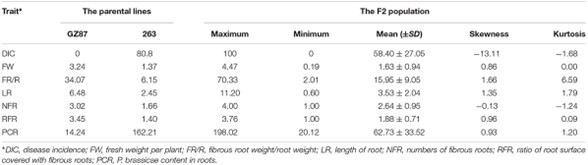
TABLE 2. Correlation among the clubroot resistance associated traits in Brassica oleracea.
2. SNP Detection and Genetic Map Construction
Out of 52,157 SNPs, 21,646 were aligned with the B. oleracea genome, with highest density on chromosome C03. A genetic linkage map was constructed with 565 bins across nine linkage groups, showing good alignment between genetic and physical positions.
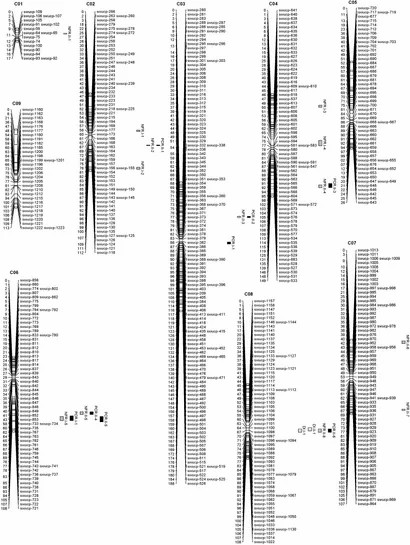 FIGURE 1. The Brassica oleracea genetic linkage map and quantitative trait loci (QTLs) for disease incidence (DIC), the number of fibrous roots (NFRs), and the P. brassicae content in roots (PCR).
FIGURE 1. The Brassica oleracea genetic linkage map and quantitative trait loci (QTLs) for disease incidence (DIC), the number of fibrous roots (NFRs), and the P. brassicae content in roots (PCR).
3. QTLs for CR-Associated Traits
Significant QTLs for disease incidence, the number of fibrous roots, and Plasmodiophora brassicae content were identified, with overlaps found between several QTLs and high R2 values for certain intervals on chromosomes C08 and C06.
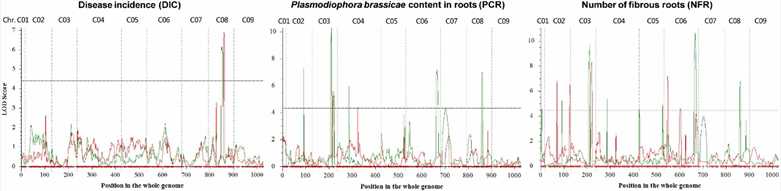 FIGURE 2. Quantitative trait loci scanning for clubroot resistance associated traits in B. oleracea in the first (red) and the second replication (green).
FIGURE 2. Quantitative trait loci scanning for clubroot resistance associated traits in B. oleracea in the first (red) and the second replication (green).
4. Comparative Analysis Among CR Loci
Certain QTLs on B. oleracea chromosomes were syntenic with known CR loci in B. rapa, although key regions on chromosomes C08 and C06 did not align with previously reported CR loci.
Conclusion
Traditional QTL methods were less efficient, but the 60K Brassica SNP microarray enabled precise mapping of B. oleracea's genetic traits. Disease incidence and root traits were used to gauge resistance, revealing new QTLs and suggesting novel resistance loci with minimal overlap with known regions.
Reference
- Peng L, Zhou L, Li Q, Wei D, Ren X, Song H, Mei J, Si J, Qian W. Identification of quantitative trait loci for clubroot resistance in Brassica oleracea with the use of Brassica SNP microarray. Frontiers in Plant Science. 2018, 18;9:822.
Here are some publications that have been successfully published using our services or other related services:
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Journal: Plant Stress
Year: 2024
The Restriction-Modification Systems of Clostridium carboxidivorans P7
Journal: Microorganisms
Year: 2023
In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Journal: Molecular Phylogenetics and Evolution
Year: 2023
Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Journal: Front. Psychiatry
Year: 2021
A high-density genetic linkage map and QTL identification for growth traits in dusky kob (Argyrosomus japonicus)
Journal: Aquaculture
Year: 2024
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium
Journal: Evolution
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
