Introduction to Ultra Low-Pass Whole Genome Sequencing
Whole genome sequencing (WGS) is a high-throughput sequencing technique that allows for a comprehensive analysis of an organism's genome, revealing its genetic variations and functional information. With the rapid advancements in sequencing technologies and the drastic reduction in associated costs, WGS has emerged as an essential tool in genomics research. However, traditional WGS often demands high sequencing depths, such as 30X or higher, which can be costly and unnecessary in certain contexts.
To address the high costs and complexity of conventional WGS, ultra-low pass whole genome sequencing (ULP-WGS) has been developed. ULP-WGS typically involves genome sequencing methods at depths below 1X. By reducing the sequencing depth, this approach decreases the data volume and costs, while employing statistical imputation and reference panels for genotype imputation and variant detection.
Technical Principles
The fundamental principle of ULP-WGS involves performing low-depth sequencing on samples, followed by using statistical methods, such as imputation, to infer the missing genotype information. This approach relies on known reference panels or population data to enhance the accuracy of genotype predictions. For instance, using reference panels allows ULP-WGS to achieve genotyping accuracy comparable to traditional WGS, even at lower sequencing depths.
Prospects for Large-Scale Genomic Research
- Large-Scale Population Studies
ULP-WGS offers significant advantages in large-scale population studies. Owing to its cost-effectiveness and efficiency, ULP-WGS can be applied in extensive cohort studies to uncover the genetic basis of complex traits and diseases in humans. In population stratification research, ULP-WGS can improve stratification accuracy by increasing sample sizes and reducing individual sequencing depths.
- Agricultural and Ecological Research
In the domains of agriculture and ecology, ULP-WGS is also regarded as a promising technology. By employing low-depth sequencing, ULP-WGS can be utilized in genome studies of crops and animals, contributing to enhanced breeding efficiency and biodiversity conservation. In agriculture, for example, ULP-WGS can detect genetic diversity in crops, thus optimizing breeding strategies.
Although ULP-WGS faces certain challenges in clinical applications, its cost-efficiency and effectiveness make it a viable alternative. In genetic diagnostics, for instance, ULP-WGS can be employed to detect chromosomal abnormalities and single nucleotide variants, supporting precision medicine initiatives.
ULP-WGS, as an emerging genomic sequencing technology, offers novel possibilities for large-scale genomic research by reducing sequencing depth and costs. Its prospects are vast in fields such as disease research, population genetics, agriculture, and ecology, indicating that it may become a pivotal tool in future genomics research.
Definition of ULP-WGS
Ultra Low-Pass Whole Genome Sequencing represents a unique form of whole genome sequencing characterized by significantly lower sequencing depths compared to traditional high-throughput methods. Its hallmark is the detection of genomic variants-such as single nucleotide polymorphisms (SNPs), copy number variations (CNVs), and structural variations (SVs)-at sequencing depths generally ranging from 0.1X to 1X.
Definition of Ultra Low-Pass Whole Genome Sequencing
ULP-WGS is defined by its utilization of extremely low sequencing depths, typically below 1X, for the identification of genomic variants. ULP-WGS is capable of detecting rare variants with moderate population frequencies (MAF >1%), but has limited sensitivity for ultra-rare or de novo mutations.
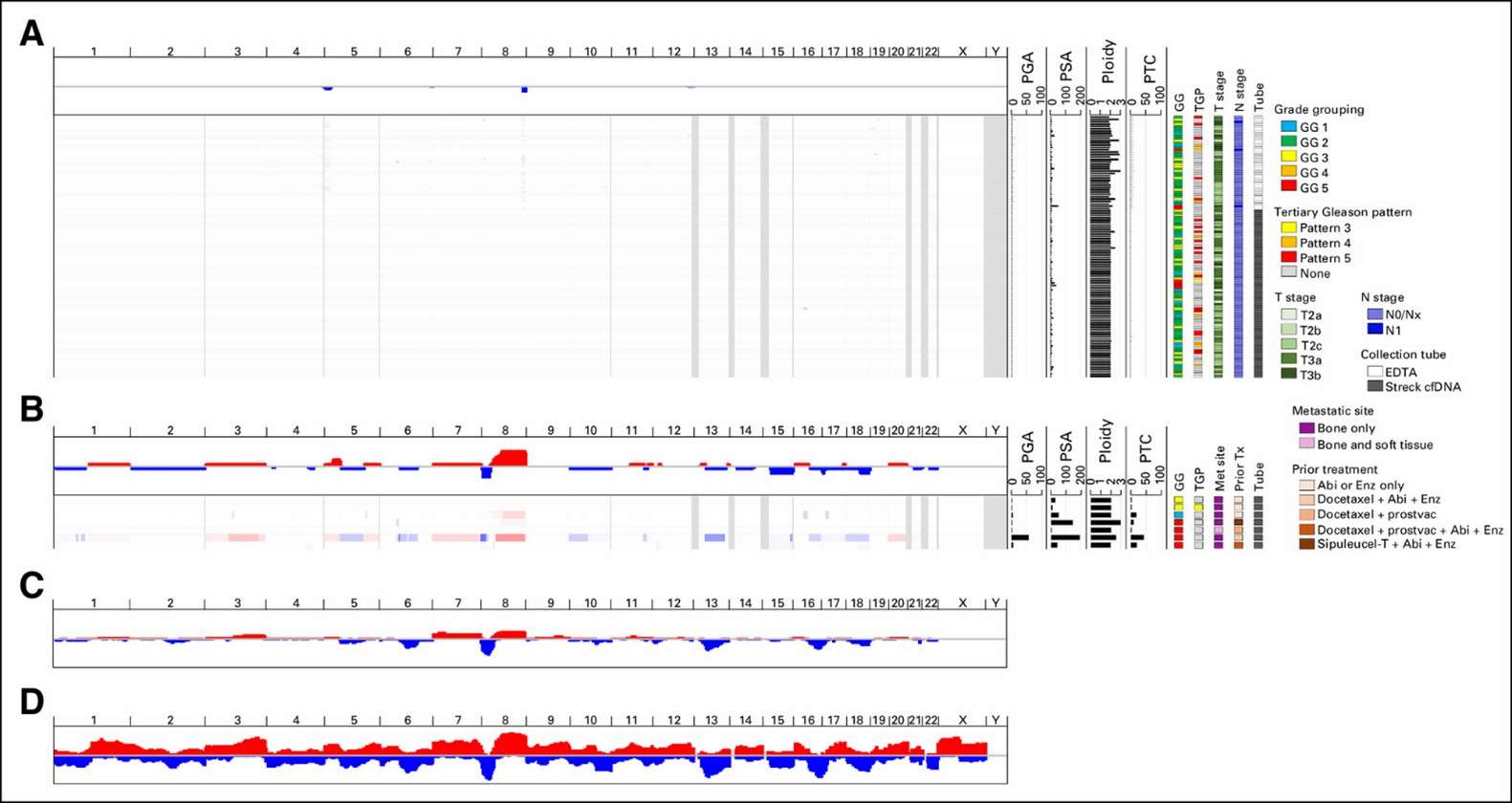 Ultra-low-pass whole-genome sequencing of circulating tumor DNA. (Hennigan, S. Thomas, et al., 2019)
Ultra-low-pass whole-genome sequencing of circulating tumor DNA. (Hennigan, S. Thomas, et al., 2019)
Comparison with Low-Pass Whole Genome Sequencing
The primary distinctions between Low-Pass Whole Genome Sequencing (LP-WGS) and ULP-WGS lie in the sequencing depth and the corresponding application scenarios:
1. Sequencing Depth:
- Low-Pass Whole Genome Sequencing: Generally has a sequencing depth between 0.5X and 1X, providing relatively high sensitivity for variant detection and a low false positive rate.
- Ultra Low-Pass Whole Genome Sequencing: Features even lower sequencing depths, typically ranging from 0.1X to 0.5X, making it suitable for scenarios where high sensitivity for variant detection is not paramount.
2. Application Scenarios:
- Low-Pass Whole Genome Sequencing: Appropriate for applications requiring high sensitivity in variant detection, such as genome-wide association studies (GWAS), genetic research of complex diseases, and rare disease diagnostics.
- Ultra Low-Pass Whole Genome Sequencing: Fits contexts where cost sensitivity and lower requirements for variant detection prevail, such as tumor content and quality assessments in cancer research, genetic studies of minority populations, and risk score computations for complex diseases.
3. Cost and Efficiency:
- Low-Pass Whole Genome Sequencing: Though relatively costly, it delivers more comprehensive results for variant detection.
- Ultra Low-Pass Whole Genome Sequencing: Offers lower costs, ideal for research institutions or regions with constrained resources.
4. Data Quality and Analysis:
- Low-Pass Whole Genome Sequencing: Provides high-quality data, suitable for complex bioinformatic analyses like structural variation mapping and internal variant profile inference.
- Ultra Low-Pass Whole Genome Sequencing: While data quality is lower, accuracy in variant detection can be enhanced through specific analytical methods, such as locus development based on extensive read evidence.
Comparative Overview: Low-Pass vs. Ultra Low-Pass Whole Genome Sequencing
| Aspect |
Low-Pass Whole Genome Sequencing |
Ultra Low-Pass Whole Genome Sequencing |
| Sequencing Depth |
0.5X-1X, high sensitivity, low false positive rate |
0.1X-0.5X, suitable for scenarios with lower variant detection requirements |
| Application Scenarios |
GWAS, complex disease studies, rare disease diagnosis |
Tumor evaluation, minority population studies, disease risk scoring |
| Cost and Efficiency |
Higher cost, more comprehensive variant detection |
Lower cost, suitable for resource-limited research |
| Data Quality and Analysis |
High quality, suitable for complex analyses (e.g., structural variation detection) |
Lower quality, but accuracy can be improved with specific analytic methods |
In summary, ULP-WGS offers a cost-effective sequencing approach tailored for specific applications, whereas LPWGS excels in sensitivity and data quality for variant detection. Choosing between these technologies depends on the specific research requirements and available resources.
Services you may interested in
Technical Principles and Advantages of ULP-WGS
Ultra-low pass whole genome sequencing provides an efficient, cost-effective approach to genomic analysis by reducing sequencing depth and optimizing data processing methodologies. This technique is particularly advantageous for analyzing large sample sets, especially when budget constraints and sample sizes are significant considerations. ULP-WGS not only detects various types of genetic variants but also significantly supports genetic research into complex diseases.
Technical Principles
- Sequencing Depth: ULP-WGS reduces sequencing depth to between 0.4 and 1.0×, thereby lowering costs while inferring missing sequences via coalescent patterns of existing genetic variants. ULP-WGS relies on population-specific reference panels (e.g., 1000 Genomes Project) rather than a single reference genome for imputation, enabling variant detection even in regions absent from reference assemblies.
- Data Processing: Utilization of tools such as GATK for sequence read processing, variant identification, and quality control is integral. Imputation analysis is employed to fill in missing sequences.
- Variant Detection: ULP-WGS is capable of detecting low-frequency, high-impact variants with statistical validity in adequately large sample sizes. Additionally, it identifies CNVs, SVs, and small nucleotide variations including SNVs and small insertions/deletions (Indels).
Advantages in Large-Scale Sample Analysis
- Cost-Effectiveness: ULP-WGS presents a high-throughput, cost-effective, and reliable alternative, particularly suitable for lower and middle-income countries or regions. Compared to traditional chromosomal microarray analysis, ULP-WGS delivers comparable information at reduced costs.
- Flexibility: This technique applies to various organisms and sample types, including complex mixed samples. It allows genotype inference without reliance on a reference genome.
- Large-Scale Sample Analysis: ULP-WGS is excellently suited for large-scale sample analysis due to its ability to provide high-quality data at reduced sequencing depths. This capability is crucial in population genetics studies and can be instrumental in assessing the genetic contribution to quantitatively measured human disease traits.
- Polygenic Risk Scoring: When combined with genotype attribution analysis, ULP-WGS facilitates polygenic risk scoring computations. This method is significant in the study of complex diseases and trait genetics.
- Broad Applicability: Beyond human genome research, ULP-WGS finds applications in agriculture, ecology, and conservation biology. In agriculture, ULP-WGS is used for evaluating genetic diversity in crops, optimizing breeding strategies, and improving trait selection efficiency.
In summary, ULP-WGS serves as a robust, flexible, and economically viable technique for diverse genomic research applications, broadening the scope and depth of genetic exploitation across various fields.
Applications of ULP-WGS in Large-Scale Genomic Research
Ultra Low-Pass Whole Genome Sequencing holds significant and broad implications in large-scale genomic research. This discussion focuses on two primary areas: its applications in population genomics and its role in elucidating the relationship between genetic variants and diseases.
1. Applications of ULP-WGS in Population Genomics
ULP-WGS technology exhibits substantial value in population genomics, particularly in the genetic analysis of complex traits and population genetics studies. Here are some specific applications:
- Genetic Analysis of Complex Traits: ULP-WGS allows for effective GWAS by capturing genomic regions of specific species. For instance, in murine models, researchers have successfully utilized ULP-WGS to achieve approximately 20% resolution across 156 unique quantitative trait loci, providing new insights into the genetic analysis of complex traits.
- Population Genetics Studies: ULP-WGS is instrumental in inferring ancestral haplotype structures and, when integrated with genotype array data, reveals population genetic architecture and evolutionary history. For example, ULP-WGS applied to individuals of European and African ancestry significantly enhances the statistical power of GWAS and minimizes errors in polygenic risk scoring.
- Detection of Rare Variants: ULP-WGS is capable of identifying rare genetic variants that may be overlooked by traditional high-throughput sequencing. In breast cancer susceptibility gene studies, it has been used to detect rare disease-associated variants.
2. Role of ULP-WGS in Elucidating Gene Variants and Disease Relationships
ULP-WGS plays a crucial role in uncovering the relationships between genetic variants and diseases, especially in the diagnosis and research of rare and complex diseases. Specific applications include:
- Diagnosis of Rare Diseases: ULP-WGS can identify gene variants associated with rare diseases. In studying neurodevelopmental disorders, for example, it has been utilized to detect small CNVs linked to various neurological conditions.
- Cancer Research: It identifies genetic mutations in cancer cells, providing critical information for cancer diagnosis, classification, prognosis, and the selection of personalized treatment plans. Analyzing ULP-WGS data from cancer patients enables researchers to discover mutation sites related to cancer progression.
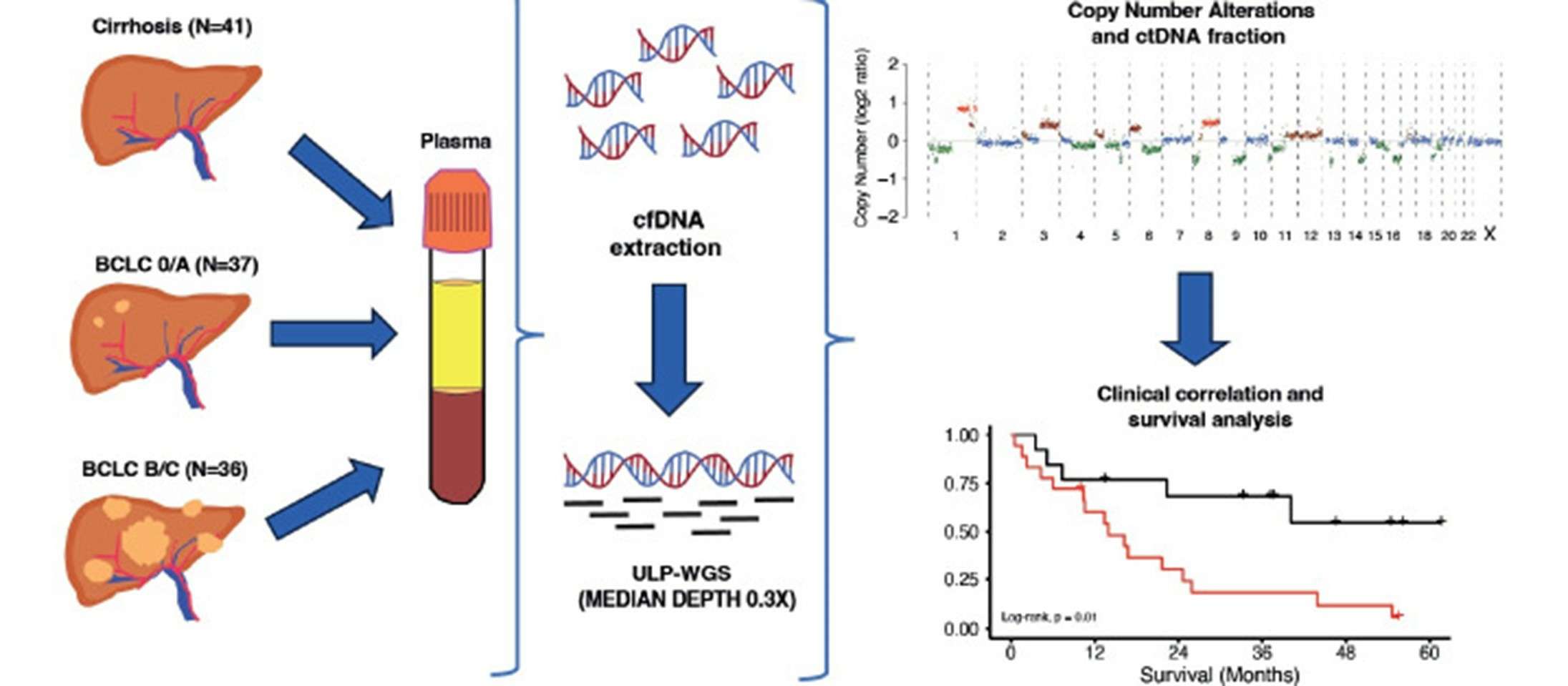 Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment. (Sogbe, Miguel, et al., 2023)
Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment. (Sogbe, Miguel, et al., 2023)
- Polygenic Risk Scoring: When combined with genotype attribution analysis, ULP-WGS is used to compute polygenic risk scores. In studies of Alzheimer's and Parkinson's diseases, ULP-WGS has demonstrated higher statistical efficiency compared to Illumina GSA in evaluating polygenic risk scores.
- Functional Annotation of Rare Variants: ULP-WGS identifies rare variants and uses functional annotation to reveal their impact on diseases. In association studies of diseases like type 1 diabetes and inflammatory bowel disease, ULP-WGS has been deployed to discover disease-linked rare variants.
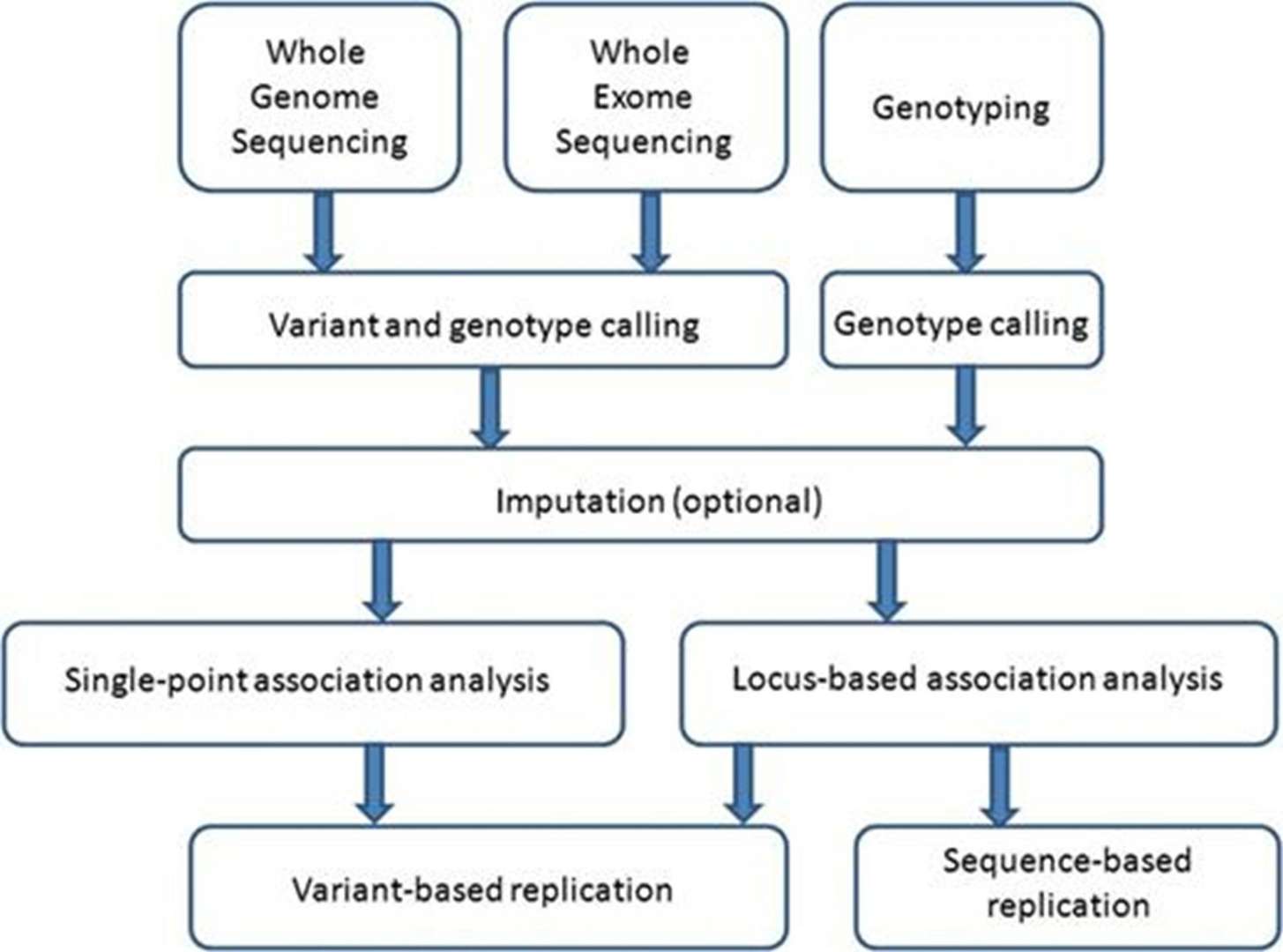 An overview of steps taken in the search for low-frequency and rare variants affecting complex traits. (Panoutsopoulou, et al., 2013)
An overview of steps taken in the search for low-frequency and rare variants affecting complex traits. (Panoutsopoulou, et al., 2013)
ULP-WGS presents extensive potential in large-scale genomic research, enhancing the precision of genetic analyses of complex traits and elucidating the link between rare variants and diseases. This technology opens new avenues for the diagnosis of rare diseases and the treatment of complex conditions. As sequencing costs continue to decrease and technology advances, ULP-WGS is set to play an increasingly prominent role in future genomic research.
Conclusion
Ultra-Low Pass Whole Genome Sequencing has emerged as a revolutionary tool in large-scale genomic research, owing to its cost-effectiveness, high throughput, and broad applicability. By reducing sequencing depth to typically 0.1-1X, ULP-WGS significantly cuts sequencing costs, making it an ideal choice for GWAS, investigations into complex diseases, and cancer genomics. This technique effectively detects SNVs, CNVs, and SVs, and in certain contexts, it can rival the performance of higher-depth sequencing. Furthermore, by leveraging extensive reference panels and sophisticated data analysis methods, ULP-WGS enhances the statistical power of genetic studies and minimizes errors in polygenic risk scoring. Current limitations include low sensitivity for small indels (<50bp) and dependency on ancestry-matched reference panels. Emerging solutions like long-read scaffolded imputation may address these gaps.
Looking forward, the evolution of ULP-WGS will depend on the optimization of sequencing depth, the integration of multi-omics data, the extension of its applications in personalized medicine, and the development of more efficient data management and analysis tools. Additionally, fostering international collaboration, building standardized protocols, and investing in skill development will be crucial for expanding the breadth and depth of its applications. Such efforts will position ULP-WGS to significantly advance genomics research and precision medicine globally, providing cost-effective solutions for disease prevention, diagnosis, and treatment, and thereby enriching the landscape of genomic science and healthcare.
References:
- Hennigan, S. Thomas, et al. "Low abundance of circulating tumor DNA in localized prostate cancer." JCO precision oncology 3 (2019): 1-13. https://doi.org/10.1200/PO.19.00176
- Gilly, Arthur, et al. "Very low-depth whole-genome sequencing in complex trait association studies." Bioinformatics 35.15 (2019): 2555-2561. https://doi.org/10.1093/bioinformatics/bty1032
- Li, Jeremiah H., et al. "Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays." Genome research 31.4 (2021): 529-537. https://www.genome.org/cgi/doi/10.1101/gr.266486.120.
- Nicod, J., Davies, R., Cai, N. et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat Genet 48, 912–918 (2016). https://doi.org/10.1038/ng.3595
- Sogbe, Miguel, et al. "Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment." Clinical and Molecular Hepatology 30.2 (2023): 177. https://doi.org/10.1093/carcin/bgae073
- Fu, J., Guo, W., Yan, C. et al. Combining targeted sequencing and ultra-low-pass whole-genome sequencing for accurate somatic copy number alteration detection. Funct Integr Genomics 21, 161–169 (2021). https://doi.org/10.1007/s10142-021-00767-y
- Panoutsopoulou, Kalliope, Ioanna Tachmazidou, and Eleftheria Zeggini. "In search of low-frequency and rare variants affecting complex traits." Human molecular genetics 22.R1 (2013): R16-R21. https://doi.org/10.1093/hmg/ddt376


 Sample Submission Guidelines
Sample Submission Guidelines
 Ultra-low-pass whole-genome sequencing of circulating tumor DNA. (Hennigan, S. Thomas, et al., 2019)
Ultra-low-pass whole-genome sequencing of circulating tumor DNA. (Hennigan, S. Thomas, et al., 2019) Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment. (Sogbe, Miguel, et al., 2023)
Prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma under systemic treatment. (Sogbe, Miguel, et al., 2023) An overview of steps taken in the search for low-frequency and rare variants affecting complex traits. (Panoutsopoulou, et al., 2013)
An overview of steps taken in the search for low-frequency and rare variants affecting complex traits. (Panoutsopoulou, et al., 2013)