Genome-wide association studies (GWAS) uncover the genetic basis of complex traits and diseases by analyzing associations between genomic variations and phenotypic characteristics. GWAS databases serve as essential resources, integrating genetic and phenotypic data to support large-scale genomic analyses. This article summarizes the features, data types, and access requirements of commonly used GWAS databases, such as GWAS Catalog, OpenGWAS, and UK Biobank. It also provides detailed guidance on data downloading and usage methods, enabling researchers to efficiently utilize these resources to advance genetic research.
What is a GWAS Database?
A GWAS database serves as a repository for genetic and phenotypic data collected from various studies, allowing researchers to access comprehensive datasets for analysis. These databases typically include genotype data (e.g., SNPs, insertions) and phenotypic data (observable traits), facilitating large-scale analyses that enhance the power to detect genetic associations.
The GWAS data we often talk about generally refers to the GWAS summary, which records the information of SNP sites one by one, including sites, P-values and other information.
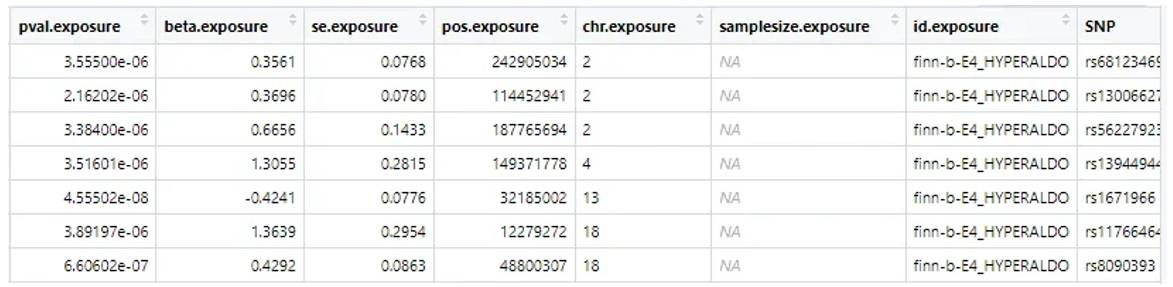 Figure 1.GWAS summary data.
Figure 1.GWAS summary data.
List of GWAS Databases
The following are some commonly used GWAS databases:
GWAS Catalog
Maintained by the European Bioinformatics Institute (EMBL-EBI), this database compiles and organizes results from genome-wide association studies (GWAS) conducted worldwide. It offers comprehensive gene-phenotype association data, including the locations of genetic variants, associated phenotypes, effect sizes, and statistical significance. The database is fully open-access, allowing users to freely search and download data.
OpenGWAS
The database, maintained by the MRC Integrative Epidemiology Unit at the University of Bristol, offers extensive data download options and API access, including some unpublished GWAS results. Researchers using this resource should regularly check for updates to access the latest data, carefully review the terms of use and citation requirements, and pay attention to quality control information and the population sources of the data.
GIANT (Genetic Investigation of ANthropometric Traits)
An international collaborative project focused on studying the genetic factors influencing human body size and shape, such as height, body mass index (BMI), and obesity levels.
UK Biobank
A large-scale database of biological samples and health information commonly used in GWAS research. It provides extensive genetic and phenotypic data suitable for Mendelian randomization analyses related to various health and disease outcomes. Accessing the data requires an application, ethical review, and payment.
EGA (European Genome-Phenome Archive)
A database specifically designed for storing and distributing genetic and phenotypic data related to biomedical research. It emphasizes privacy and data security, especially for sensitive human genetic information. EGA contains data from GWAS, sequencing projects, and other large-scale genetic studies linked to disease and normal phenotypic traits. Accessing EGA data involves a stringent process, requiring researchers to submit an application detailing the purpose of use, which must be approved before data access is granted.
dbGaP (Database of Genotypes and Phenotypes)
Managed by the National Center for Biotechnology Information (NCBI), this database archives, curates, and disseminates research data exploring the interaction between genotypes and phenotypes. The data typically originate from GWAS, medical sequencing projects, molecular diagnostic studies, and other genetic and clinical research fields. Access requires a detailed application process, including submission of a research plan, purpose of use, and often approval from an Institutional Review Board (IRB) or ethics committee. Approved researchers gain access to specific datasets for their projects.
FinnGen
A Finnish genetic database containing extensive genetic data from the Finnish population. Due to the population's high genetic homogeneity, it is particularly valuable for identifying the genetic basis of complex diseases. The data is publicly available for research purposes, but access may require adherence to specific policies or collaboration agreements.
IEU (Instrumental Variables Estimation in Epidemiology Unit)
This database and resource platform provide instrumental variables (typically single nucleotide polymorphisms, SNPs) and methodological support for Mendelian randomization analyses. Many of the methodological resources and some instrumental variable data are freely available.
PGC (Psychiatric Genomics Consortium)
A consortium focused on genetic research related to psychiatric disorders. It provides GWAS data that are critical for studying causal mechanisms underlying mental health issues.
CKD Gen
A database specializing in the genetic study of chronic kidney disease and related traits, such as estimated glomerular filtration rate (eGFR). It supports Mendelian randomization analyses in kidney disease research.
GEFOS (Genetic Factors for Osteoporosis)
An alliance focused on the genetics of osteoporosis, providing GWAS data related to bone health and osteoporosis.
GLGC (Global Lipids Genetics Consortium)
Offers genetic data related to blood lipid levels, serving as a valuable resource for Mendelian randomization studies in cardiovascular and related fields.
SSGAC (Social Science Genetic Association Consortium)
Focuses on genetic research in social science areas such as socioeconomic status and educational attainment.
Data generated by consortia (e.g., PGC, CKD Gen, GEFOS, GLGC, SSGAC) or projects are often shared freely with the research community through scientific publications or public databases once published.
How to Download GWAS Data
Search papers in pubmed
Literature Search: Begin by searching PubMed using keywords such as "GWAS," "genome-wide association study," along with the name of the specific disease or trait of interest. This will help identify relevant research articles.
Abstract Review: Reviewing the abstracts can provide an initial assessment of the relevance of the studies and the applicability of the data.
Identifying Data Sources: Many GWAS studies mention the datasets they used or specify the public databases where the data is stored, such as the GWAS Catalog, EGA (European Genome-Phenome Archive), or dbGaP (Database of Genotypes and Phenotypes).
Accessing GWAS Databases and Resources: Following the guidance in the literature, directly access databases like the GWAS Catalog. These databases often provide data download options, which may require account registration and agreement to specific usage terms.
Download in OpenGWAS database
After entering the official website, the interface is as follows,Do a search in the search bar, type in the name of a disease or trait, for example "fat".
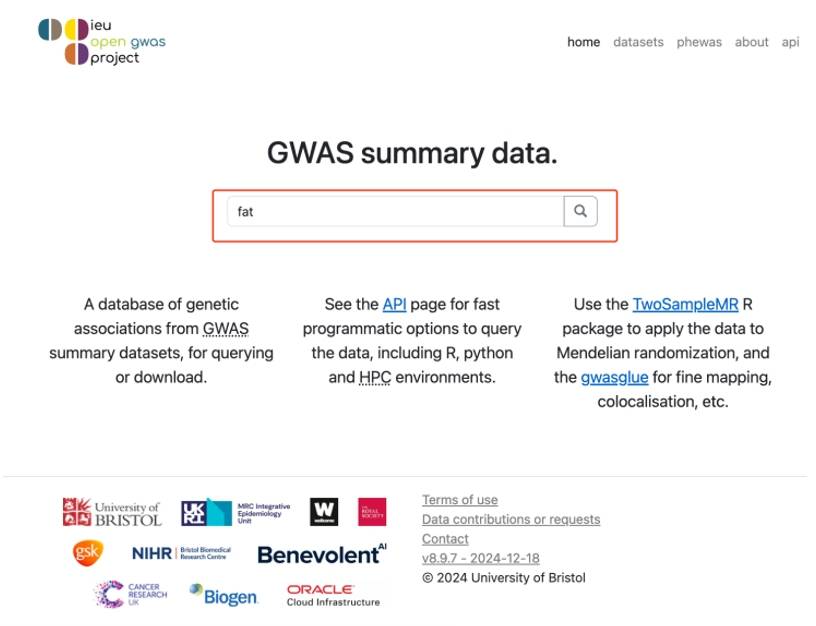 Figure 2. OpenGWAS database home page.
Figure 2. OpenGWAS database home page.
The search results are some basic information, mainly looking at the ID, because we can use this ID to directly analyze the TwoSampleMR package, the first alphabet of the ID to tell which database it belongs to.
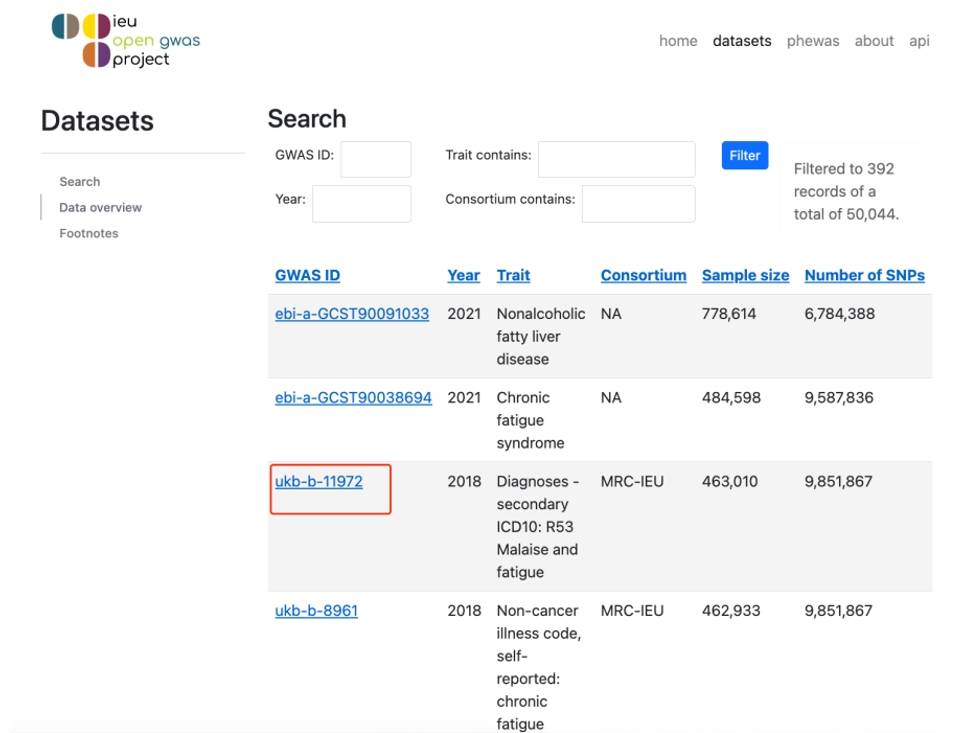 Figure 3. OpenGWAS database search page.
Figure 3. OpenGWAS database search page.
Click the data in the red box, enter to get a detailed description of the data, if you see this data set has a Download VCF flag, this data is generally downloadable, generally speaking, if there is no such flag, it indicates that the data may not be open or can not be downloaded, but this is not absolute.
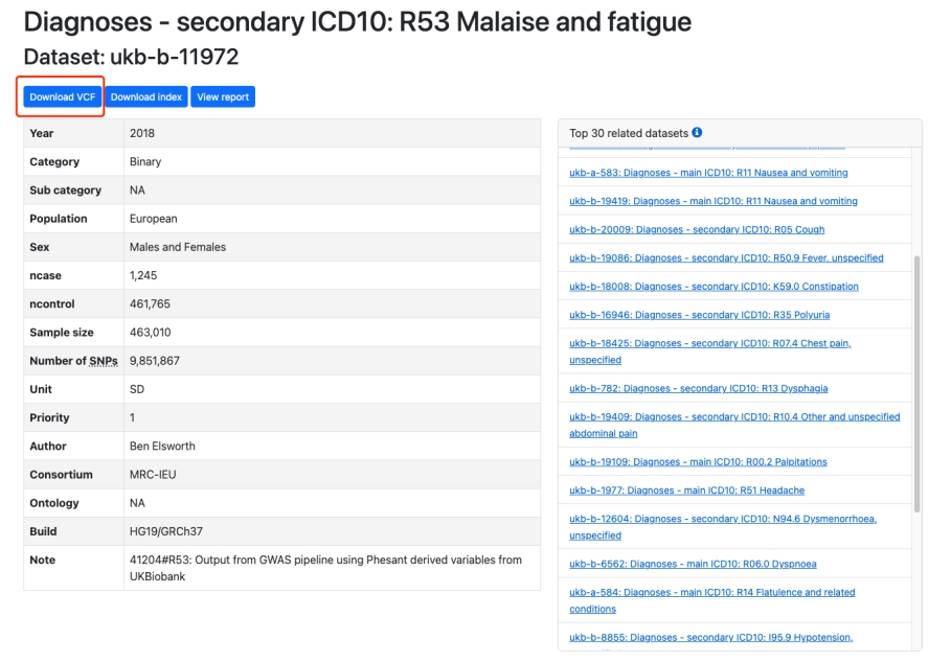 Figure 4. OpenGWAS database download page.
Figure 4. OpenGWAS database download page.
Download in GWAS Catalog database
This is a commonly used database, but this database often has some incomplete data, so we need to pay attention to when searching.Go to the next screen from Summary statistics.
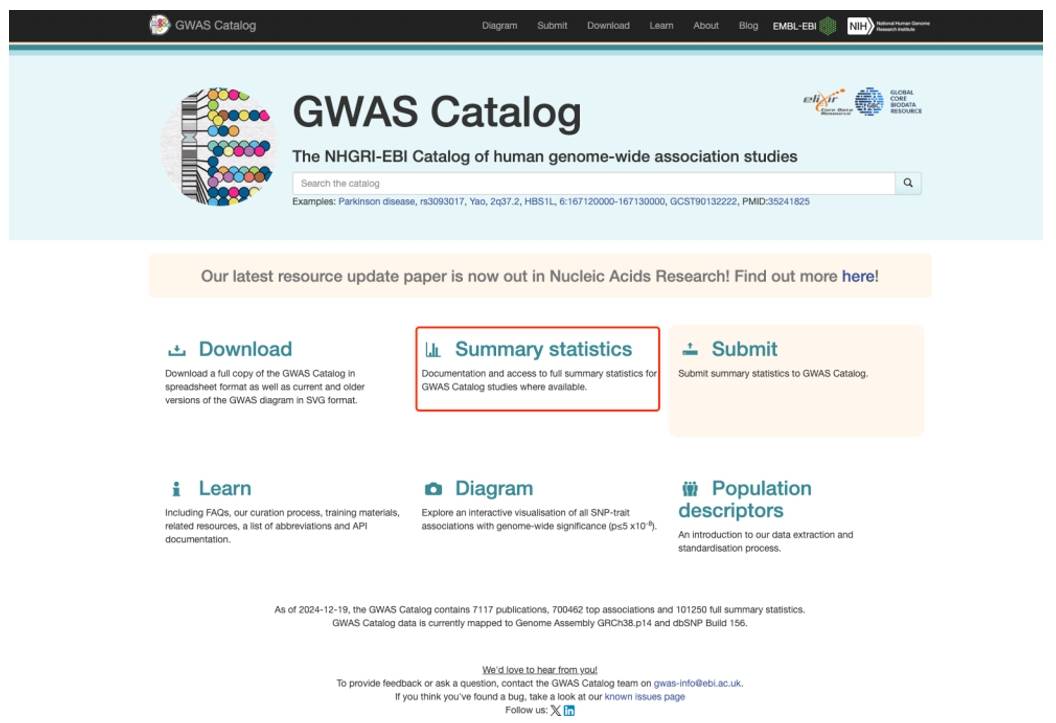 Figure 5. GWAS Catalog database home page.
Figure 5. GWAS Catalog database home page.
Go to the next search screen from Available studies.
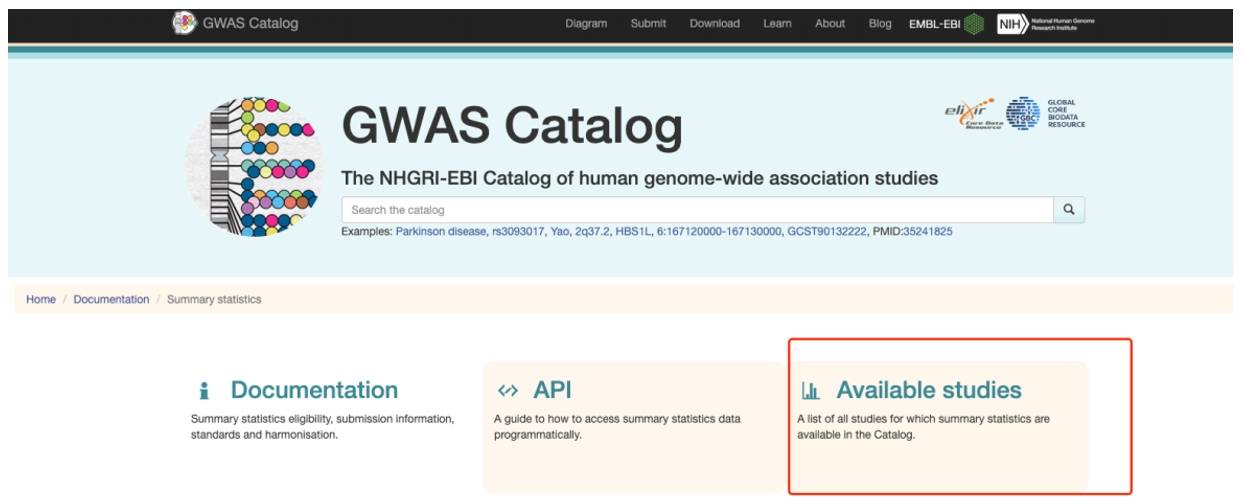 Figure 6. GWAS Catalog database summary statistics page.
Figure 6. GWAS Catalog database summary statistics page.
The Traits option allows users to search for and locate the required data. When accessing downloadable files via FTP Download, it is important to check the file size; files around 400 MB or larger typically contain complete datasets, while smaller files may lack sufficient SNP loci for GWAS analysis. Additionally, some datasets with GCST-prefixed IDs can also be found in the openGWAS database.
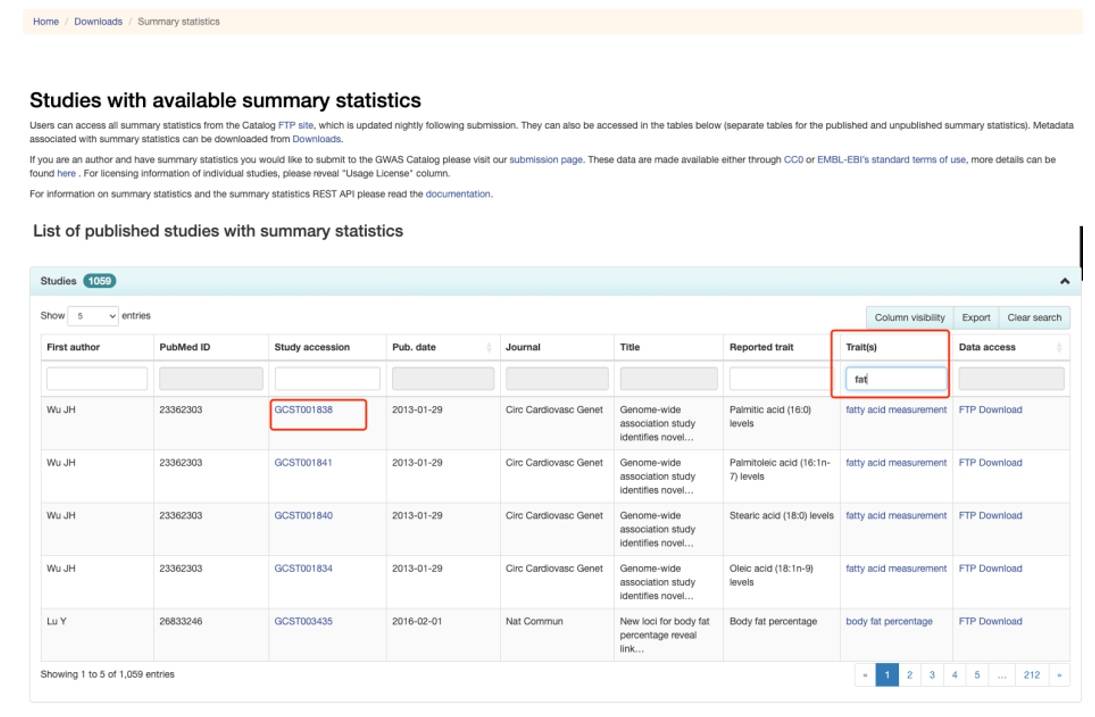 Figure 7. GWAS Catalog database search page.
Figure 7. GWAS Catalog database search page.
In conclusion, GWAS databases play a pivotal role in advancing genetic research by providing essential resources that facilitate the exploration of genetic factors underlying complex traits and diseases. With a wide range of publicly accessible and specialized databases such as the GWAS Catalog, OpenGWAS, and UK Biobank, researchers can efficiently access vast amounts of genetic and phenotypic data.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1.GWAS summary data.
Figure 1.GWAS summary data. Figure 2. OpenGWAS database home page.
Figure 2. OpenGWAS database home page. Figure 3. OpenGWAS database search page.
Figure 3. OpenGWAS database search page. Figure 4. OpenGWAS database download page.
Figure 4. OpenGWAS database download page. Figure 5. GWAS Catalog database home page.
Figure 5. GWAS Catalog database home page. Figure 6. GWAS Catalog database summary statistics page.
Figure 6. GWAS Catalog database summary statistics page. Figure 7. GWAS Catalog database search page.
Figure 7. GWAS Catalog database search page.