Mitochondria, found in almost all cells, are organelles similar to miniature power stations that generate the energy needed to maintain basic cell functions. The genetic information of mitochondria is stored in mitochondrial DNA (mtDNA), which is mostly in the form of circular molecules, but also includes linear molecules. Unlike nuclear DNA, mtDNA is a naked double-stranded DNA molecule that is not protected by histones and is located near reactive oxygen species. These factors, coupled with the relatively low fidelity of mitochondrial DNA replication and repair mechanisms, lead to a higher mutation rate than nuclear DNA. Over time, this high mutation rate has made mtDNA an important indicator for studying the effects of environmental stress and other damaging factors. Extracting mitochondrial DNA can not only help us deeply explore the pathogenesis of mitochondrial diseases and provide key clues for developing targeted treatment methods, but also play an important role in the fields of forensic medicine, anthropology and other fields.
Service you may interested in
Resources
Traditional mitochondrial DNA isolation methods
1)Cesium chloride ultracentcularization
Cesium Chloride Ultracentitation is a classic and efficient method for extracting high-purity mitochondrial DNA (mtDNA). This technology relies on the principle of cesium chloride density gradient centrifugation and can effectively separate DNA molecules of different densities. The following are the detailed steps after optimization, describing how to use the cesium chloride ultracentury to extract mtDNA:
Materials and reagents
Cesium chloride (CsCl)
Ethidium Bromide (EtBr): Used for staining DNA for easy observation under ultraviolet light
centrifuge tube
ultracentrifuge
ultraviolet lamp
Appropriate amount of buffer (usually PBS or TE buffer)
Enzyme digestion reagent (optional, for digestion of non-mtDNA)
Extraction steps
Cell lysis: Place cells or tissue samples in suitable lysis buffer. Use mechanical or chemical methods (such as ultrasound, freeze-thaw cycles, or the use of lysing agents) to destroy cell membranes and release mitochondria.
Preliminary centrifugation: Remove cell debris and undisrupted cells by low-speed centrifugation (approximately 1000 g, 10 minutes).
The supernatant was collected, which contained mitochondria.
Mitochondrial preparation: The supernatant was centrifuged at high speed (approximately 10000 g, 15 minutes) to precipitate mitochondria.
Discard the supernatant and resuspend the mitochondrial pellet in a small amount of PBS or TE buffer.
Adding cesium chloride and dye: Add cesium chloride and a small amount of ethidium bromide (EtBr) to the mitochondrial suspension. Ethidium bromide binds to DNA, making the DNA visible under ultraviolet light. Adjust the density of the mixture to ensure that the DNA is correctly layered in the cesium chloride density gradient.
Ultracentularation: Transfer the mixture into ultracentular-centrifuge tubes and centrifuge (about 100,000 g or more, which usually takes several hours to overnight). During centrifugation, DNA will be layered based on its density, and mitochondrial DNA (mtDNA) and nuclear DNA are separated due to different densities.
DNA extraction: Use an ultraviolet light to inspect the centrifuge tube and locate the glowing DNA band. Carefully aspirate bands containing mtDNA (usually located in lighter locations because mtDNA is smaller than nuclear DNA). Transfer the extracted mtDNA into a new container.
DNA purification: Use alcohol precipitation or other DNA purification methods to remove cesium chloride and ethidium bromide. Resupend the DNA in TE buffer or other suitable solution.
2)Column chromatography
Extraction of mitochondrial DNA (mtDNA) by column chromatography is an effective biomolecule separation technique that relies on the interaction between a stationary phase (column material) and a mobile phase (buffer) to separate different biomolecules. This method is particularly useful when extracting mtDNA because it reduces contamination of nuclear DNA and improves mtDNA purity. The following are the steps for extracting mtDNA using column chromatography, including the required materials and operating details.
Materials and reagents
Lysis buffer: Usually contains non-ionic surfactants to destroy cell membranes.
Column chromatography column: Commonly used are silica gel matrix or ion exchange resin.
Elution buffer: pH and salt concentration are adjusted to optimize elution of DNA.
Centrifuge tube: Used to collect eluted samples.
Centrifuge: Used for sample removal and elution steps.
Microcentrifuge tube: Used to collect the final DNA solution.
Ethanol or isopropanol: Used for DNA precipitation.
TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0): Used to dissolve purified DNA.
Extraction steps
Cell lysis: Mix target cells or tissues with lysis buffer. Completely lyse cells and release mitochondria by means such as slight shaking, freeze-thaw cycles, or using ultrasound.
Mitochondrial isolation: Remove cell debris and large undisrupted cells by low-speed centrifugation (e.g., 1,000 × g, 10 minutes).
The supernatant is collected and the mitochondria are then precipitated by high-speed centrifugation (e.g., 10,000 × g, 15 minutes). The supernatant was discarded and the precipitated mitochondria were resuspended in a small amount of lysis buffer.
DNA extraction: Add appropriate amount of nucleic acid extraction buffer and protease K to the mitochondrial suspension to digest the protein and release DNA. DNA was extracted using the phenol-chloroform method, and then precipitated with ethanol or isopropanol.
Column chromatography purification: Resuspend the precipitated DNA in appropriate buffer and load it into a previously prepared column chromatography column. Elute non-specifically adsorbed substances with elution buffer. Gradually increase the salt concentration in the elution buffer to specifically elute mtDNA. Collect the eluate, which contains highly pure mtDNA.
DNA precipitation and resuspension: Add 2.5 volumes of 100% ethanol and 1/10 volumes of 3M sodium acetate to the eluate, mix, and place at-20 ° C overnight to precipitate DNA. After freezing precipitation, the DNA precipitate is collected by centrifugation (e.g., 12,000 × g, 30 minutes, 4°C). Discard the supernatant, wash the DNA precipitate at least once with 70% ethanol, and then centrifuge to recover the DNA. Air-dry or briefly vacuum dry the DNA precipitate to avoid complete drying, which makes it difficult to resuspend the DNA. The dried DNA was resuspended in a small amount of TE buffer, completely dissolved, and stored at-20 ° C.
3)DNase method
DNase extraction of nuclear DNA (ntDNA) is an effective technique that purifies nuclear DNA by using DNase I, an enzyme that specifically cleaves DNA, to digest and remove mitochondrial DNA (mtDNA) or other non-target DNA in a sample. This method is particularly suitable for experiments that require high-purity nuclear DNA, such as genomic research at the chromosome level or forensic applications. The following are the detailed steps for extracting nuclear DNA using the DNase method:
Materials and reagents
DNase I: An enzyme that specifically cleaves single and double strands of DNA.
Buffer: DNase I reaction buffer is usually used, and the specific components are prepared according to the enzyme's instructions.
Lysis buffer: Used for cell lysis and release DNA.
Proteinase K: Used for protein digestion to ensure DNA release.
Phenol chloroform solution: Used for DNA extraction and purification.
Ethanol or isopropanol: Used for DNA precipitation.
TE buffer: Used for resuspension of DNA.
Centrifuge tubes and microcentrifuge tubes: used for sample processing and collection.
Centrifuge: Used for various centrifugation steps.
Temperature-controlled water bath or heating block: Used to control the temperature of the enzyme treatment step.
Extraction steps
Cell lysis: Place a cell or tissue sample in a centrifuge tube containing lysis buffer. Proteinase K, usually at a concentration of 100 μg/mL, is added to the centrifuge tube to digest protein and release DNA. Samples were incubated at 56°C for several hours or overnight to completely lyse the cells.
DNase I processing: Prepare DNase I solution: Prepare DNase I reaction buffer according to the manufacturer's instructions and add DNase I (usually 1-10 units/μg DNA). Add DNase I solution to the lysed sample and mix well. Incubate at 37°C for an appropriate time (usually 30 minutes to 1 hour) to allow DNase I to fully cleave mtDNA. DNase I activity can be terminated by adding EDTA to a final concentration of 10 mM, or by other methods recommended in the instructions.
DNA extraction: Add an equal volume of phenol chloroform solution to the processed sample, shake it fully, and then centrifuge and layer it. Carefully transfer the DNA-containing aqueous phase supernatant into a new centrifuge tube. Repeat the phenol-chloroform extraction step if necessary to increase the purification effect.
DNA precipitation: Add 0.1 volume of 3M sodium acetate and 2.5 volumes of 100% isopropanol to the supernatant. After mixing, refrigerated at-20 ° C overnight to promote DNA precipitation. The next day, the DNA precipitate is collected by centrifugation (e.g., 12,000 × g, 30 minutes, 4°C), the supernatant is discarded, and the precipitate is washed with 70% ethanol. Air-dry or gently heat until the DNA precipitate is just dry to avoid excessive drying, and then resuspend the DNA with appropriate amount of TE buffer.
4)Alkaline lysis procedure
Alkali denaturation method is a simple, rapid and low-cost method to extract mitochondrial DNA (mtDNA), which is especially suitable for small-scale experiments and preliminary research. This method is based on the dissociation characteristics of DNA under alkaline conditions. mtDNA can be effectively released and purified through simple alkaline treatment, especially when extracted from cell culture or a small number of tissue samples. The following are the detailed steps for extracting mtDNA using alkali denaturation method:
Materials and reagents
NaOH (sodium hydroxide): The common concentration is 50 mM.
EDTA: Final concentration of 1 mM, used to chelate metal ions and prevent DNase activity.
Tris-HCl: Used to neutralize DNA solutions, pH 8.0, usually at a concentration of 1M.
Ice-cold ethanol: Used for DNA precipitation.
TE buffer: For DNA resuspension, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
Microcentrifuge tubes.
Centrifuge.
Vortex mixer.
Water bath or heating block: Used to heat samples.
Extraction steps
Sample preparation: Collect cell or tissue samples in microcentrifuge tubes. Add the appropriate amount of PBS buffer and remove excess buffer and impurities by centrifugation (3000 × g, 5 minutes).
Alkaline treatment: Add a mixture of 50 mM NaOH and 1 mM EDTA to the sample, with a volume of approximately 100-200 μL depending on the sample size. After a brief vortex mixing, the sample was heated in a 95°C water bath for 10-30 minutes to allow complete release of the DNA.
Neutralization reaction: Remove the sample from the heating block and immediately add an equal volume of 1M Tris-HCl, pH 8.0 to neutralize it.
After mixing, swirl briefly and cool quickly to room temperature.
DNA precipitation: Add 2-2.5 volumes of ice-cold ethanol to the neutralized sample and mix well.
Freeze the mixture at-20 ° C for at least 2 hours or overnight to maximize DNA precipitation.
Collect DNA: Centrifuge the sample at 12000 × g for 15 minutes at 4°C to collect the DNA precipitate. The supernatant was discarded, the DNA precipitate was washed with 70% ice-cold ethanol, and the centrifugation step was repeated. Discard the supernatant again and air-dry the DNA precipitate to avoid excessive drying.
Resuspend DNA: Resuspend the dried DNA pellet in the appropriate amount of TE buffer. Completely dissolve the DNA by gently swirling or placing it at room temperature for several hours.
Emerging mitochondrial DNA isolation technology
Magnetic bead separation method
Magnetic bead separation method is an innovative mitochondrial DNA separation technology. Its unique mechanism is based on the interaction of specific ligands modified on the surface of magnetic beads with mitochondria or mitochondrial DNA. Magnetic beads are usually composed of magnetic materials (such as magnetite or magnetic iron oxide) with a functional surface modified with ligands that can specifically bind to mitochondria or mitochondrial DNA. These ligands can be antibodies, nucleic acid aptamers, etc. Taking antibody-modified magnetic beads as an example, antibodies can specifically bind to specific proteins on the surface of mitochondria. When magnetic beads are added to the cell lysate, the antibodies on the surface of the magnetic beads recognize and bind to the target protein on the surface of the mitochondria to form a magnetic bead-mitochondrial complex. Under the action of an external magnetic field, the magnetic bead-mitochondrial complex will move towards the magnetic pole and be separated from other impurities. In this way, mitochondria can be separated from cell lysates quickly and efficiently. If mitochondrial DNA is to be isolated directly, magnetic beads modified with nucleic acid aptamers that can specifically bind to mitochondrial DNA can be used. Aptamers can bind to specific sequences of mitochondrial DNA, thereby achieving selective capture of mitochondrial DNA.
Kit-based separation method
Common mitochondrial DNA isolation kits on the market have their own working principles and operating procedures. Kits based on the principle of silica gel membrane adsorption are widely used. Taking a certain brand of mitochondrial DNA isolation kit as an example, its operating process is as follows: First, cell samples are collected and centrifuged to remove the culture medium. Then add an appropriate amount of cell lysis buffer, which contains special ingredients that can destroy cell membranes and release cytoplasm and mitochondria. Cell debris and large particulate matter are then removed by a centrifugation step, and the cell lysate is transferred to a centrifuge column containing a silica gel membrane. Under high-salt conditions, mitochondrial DNA will be specifically adsorbed on the silica gel membrane, while other impurities will flow out with the eluent. The silica gel membrane was then washed with washing buffer to remove residual impurities. Finally, the mitochondrial DNA adsorbed on the silica gel membrane was eluted with low-salt elution buffer. This kit based on silica gel membrane adsorption is relatively simple to operate, can effectively remove nuclear DNA and other pollutants, and is suitable for large-scale sample processing.
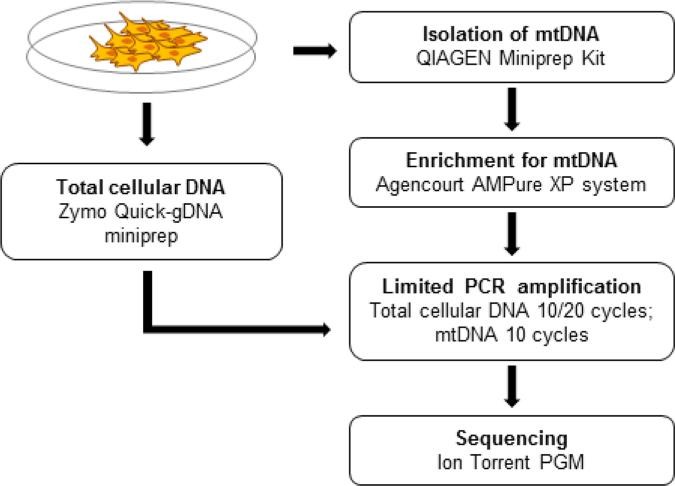 Figure 1 .Total cellular DNA is isolated for control purposes.(Quispe-Tintaya, W.,et.al,2013)
Figure 1 .Total cellular DNA is isolated for control purposes.(Quispe-Tintaya, W.,et.al,2013)
There are various methods for extracting mitochondrial DNA (mtDNA), including ultracentular-centrifugation, column chromatography, DNase treatment and alkali denaturation, each with its own advantages and disadvantages. Ultracentrifugation has high purity but complex operation, column chromatography reduces nuclear DNA contamination, DNase method improves nuclear DNA purity, and alkali denaturation method is simple but may affect integrity. As an emerging technology, magnetic bead separation method has the advantages of speed and efficiency. Choosing the right method is crucial to the accuracy of the study.
Reference:
- Quispe-Tintaya, W., White, R. R., Popov, V. N., Vijg, J., & Maslov, A. Y. (2013). Fast mitochondrial DNA isolation from mammalian cells for next-generation sequencing. BioTechniques, 55(3), 133–136. https://doi.org/10.2144/000114077


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1 .Total cellular DNA is isolated for control purposes.(Quispe-Tintaya, W.,et.al,2013)
Figure 1 .Total cellular DNA is isolated for control purposes.(Quispe-Tintaya, W.,et.al,2013)