Mutations within the mitochondrial DNA (mtDNA) sequence are frequently observed in specific types of tumors. There are two discernible categories of mtDNA variants associated with cancer: de novo mutations acting as oncogenic "inducers" and functional variants serving as "adapters," enabling cancer cells to thrive in diverse environments. These mtDNA variants originate from three sources: hereditary genetic variations that run in families, somatic mutations occurring within individual cells, and variants linked to ancient mtDNA lineages (haplogroups) renowned for their adaptability to shifting organizational or geographic conditions.
Beyond variations in the mtDNA sequence, alterations in mtDNA copy numbers and the potential transfer of mtDNA sequences to the cell nucleus may also contribute to the development of certain cancers. The functional significance of mtDNA variants has been unequivocally demonstrated in eosinophilic cell tumors and prostate cancer. Additionally, mtDNA variants have been reported across a spectrum of other cancer types.
The pivotal role of mitochondrial metabolism in cancer is substantiated by modifications in mitochondrial genes encoded by nuclear DNA, which impact the production of mitochondrial reactive oxygen species, the redox state, and mitochondrial intermediates—vital substrates for chromatin-modifying enzymes. Consequently, even subtle changes in mitochondrial genotype can exert profound effects not only on the cell nucleus but also on the processes of carcinogenesis and cancer progression.
Mitochondrial DNA (mtDNA) Variation and Its Impact on Metabolism
The primary source of cellular energy is the mitochondria, which generates adenosine triphosphate (ATP). This energy production process relies on the presence of oxygen and an oxidizable carbon source, such as glucose. As glucose undergoes metabolism to produce water and carbon dioxide, ATP is concurrently generated along with several high-energy intermediates. The production of these intermediates is closely tied to the status of the mitochondrial genome. Oncogenic mutations within mitochondrial genes can disrupt the enzymes of the tricarboxylic acid cycle, leading to alterations in mitochondrial metabolite levels.
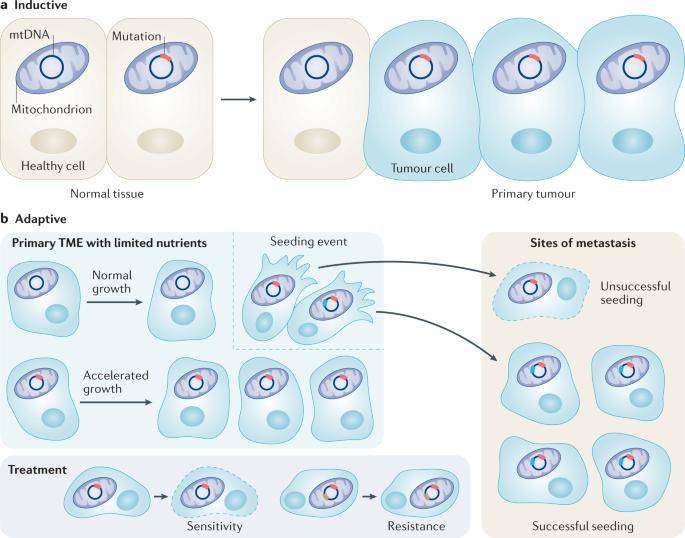 Mitochondrial DNA variants can play inductive and adaptive roles in oncogenesis. (Kopinski et al., 2021)
Mitochondrial DNA variants can play inductive and adaptive roles in oncogenesis. (Kopinski et al., 2021)
Clinically, there are three primary categories of relevant mtDNA variants.
- The first category encompasses maternally inherited variants that originate in the female germ line. Initially, these variants emerge within a cell dominated by normal mtDNA, resulting in a mixed cytoplasm containing both variant and reference mtDNA, a condition termed heterozygosity.
- A second class of clinically significant mtDNA variants arises in oocytes during development or within somatic tissues throughout an individual's lifetime. These mutations encompass base substitutions and deletions, accumulating with age and potentially contributing to the process of biological aging. Deletions can lead to a variety of symptoms, depending on their distribution within tissues and the degree of heterogeneity. Examples include Pearson's syndrome, Kearns-Sayre syndrome, and chronic progressive external oculomotor paralysis.
- The third group of clinically relevant mtDNA variants consists of ancient variants that emerged during human migration, both in Africa and around the world. Due to their ancient origin, these variants have become relatively uniform within specific populations. As people migrated to new geographical environments, advantageous functional variants were naturally selected, giving rise to distinct regional populations characterized by unique sets of mtDNA haplotypes, known as haplogroups. For instance, when humans migrated out of Africa to Eurasia and the Americas, only two primary mtDNA haplotypes left the African continent. Subsequent generations acquired additional variants as they adapted to their new surroundings.
Comprehensive mtDNA Sequencing Unraveling Cancer's Genetic Clues
Through meticulous scrutiny of Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) data derived from The Cancer Genome Atlas (TCGA), various research teams have unearthed potential correlations between mitochondrial DNA (mtDNA) variants and cancer. In essence, the primary criterion employed to ascertain the relevance of these mtDNA variants to cancer involves their presence within the tumor samples of patients as opposed to their adjacent normal tissues, as revealed by mtDNA sequencing. This approach has yielded a substantial number of candidate mtDNA variants with potential links to cancer.
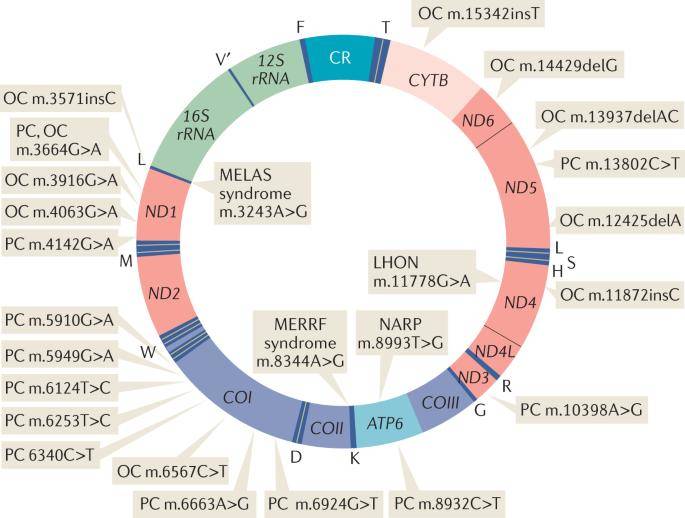 Human mtDNA map showing representative variants. (Kopinski et al., 2021)
Human mtDNA map showing representative variants. (Kopinski et al., 2021)
However, it is worth noting that the pathogenicity of these cancer-associated mtDNA variants has not been conclusively established in the majority of extensive investigational studies and numerous tumor-specific investigations, despite the insights gained from mtDNA sequencing. This lack of confirmatory evidence is, in part, attributable to the intricate relationship between mtDNA variants and their biological implications, which can vary significantly depending on factors such as tissue type, the tumor microenvironment, and nuclear DNA (nDNA) genotype.
The criteria for evaluating the pathogenicity of specific mtDNA variants encompass the need for independent, replicated observations of these mutations within the tumor environment, the demonstration of associated biochemical defects revealed through mtDNA sequencing, the transmission of biochemical effects from one cell to another through mtDNA, and the establishment of these associations without any influence from nDNA or disease-specific factors.
To date, the only mtDNA mutations in cancer that have undergone such rigorous scrutiny, including mtDNA sequencing, are those observed in cases of prostate cancer and eosinophilic tumors.
Tumor-Associated Mitochondrial DNA Mutations
Numerous studies have delved into the presence of mitochondrial DNA (mtDNA) variants within specific types of tumors. In certain instances, the cumulative load of acquired mtDNA variants serves as a potential biomarker for tumorigenicity. For instance, the detection of mtDNA mutations in cells associated with prostate cancer may contribute to an augmented tumorigenic potential. Prostate cancer-linked mtDNA mutations often co-occur with nucleosome-driven events, emphasizing the pivotal role of mtDNA mutations as co-initiators in cancer development. Additionally, there is emerging evidence suggesting that mtDNA mutations can act as tumor promoters in cystic cell carcinoma of the thyroid, breast cancer, pancreatic cancer, gynecological malignancies, lung adenocarcinoma metastases, and acute myeloid leukemia. In contrast, the role of mtDNA variants appears to be of limited significance in glioblastoma.
Mitochondrial Gene Mutations and Cancer Epigenome Regulation
The regulation of gene expression within the cell nucleus has been extensively studied and is known to involve various epigenetic modifications of DNA and histones. These modifications are closely tied to the metabolites produced by mitochondria. Histone acetylation, for instance, relies on acetyl coenzyme A, a molecule derived from citrate. This acetylation process can be reversed by NAD-dependent sirtuins. On the other hand, histone and DNA methylation are dependent on S-adenosylmethionine, which can be reversed by enzymes utilizing α-ketoglutarate (α-KG) as a substrate. Interestingly, α-KG itself can be inhibited by succinate and fumarate. Furthermore, a significant group of chromatin remodeling proteins operate in an ATP-dependent manner.
Notably, mutations in genes responsible for encoding IDH1 and IDH2, which are enzymes catalyzing the conversion of isocitrate to α-KG, have been linked to various cancers such as gliomas, acute myeloid leukemias, and several solid tumors. α-KG serves as a cofactor for α-KG-dependent dioxygenases, including the Jumonji C (JmjC) histone demethylase and the Ten-Eleven Translocation (TET) dioxygenase family, both of which play pivotal roles in DNA demethylation processes.
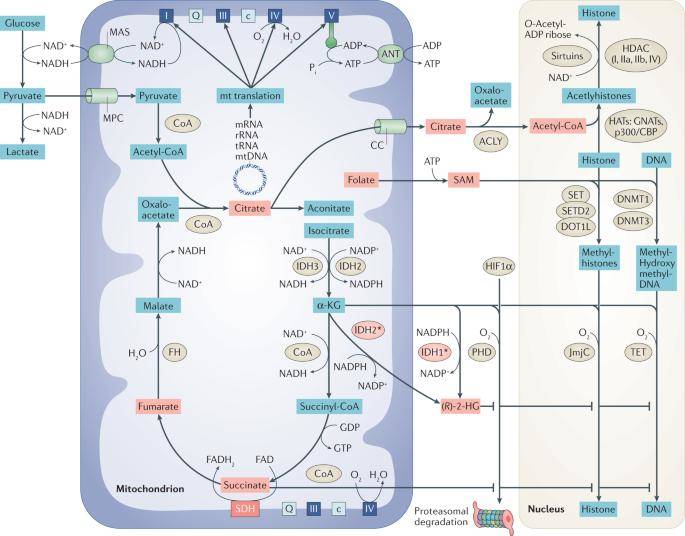 Mitochondrial biology and metabolism and its relationship to the epigenome. (Kopinski et al., 2021)
Mitochondrial biology and metabolism and its relationship to the epigenome. (Kopinski et al., 2021)
Heterogeneous mtDNA Alterations and Tumor Epigenome
This concept gains substantial support through a comparative analysis of nuclear DNA (nDNA) methylation patterns across different cell lines: osteosarcoma 143B cells, breast cancer MDA-MB-435 cells, and permanent breast epithelial MCA 12A cells, both with and without intact mtDNA. The loss of mtDNA precipitates discernible changes in CpG island methylation profiles. Remarkably, reintroducing mtDNA into these cells partially restores CpG island methylation.
Further underscoring this phenomenon, a tumor sample extracted from a patient afflicted with renal cell carcinoma exhibited the presence of the mtDNA tRNALeu(UUR) (MT-TL1) m.3243A>G, G allele mutation within the tumor tissue, with a mutation load reaching 89%. In contrast, the patient's kidneys contained approximately 50% mtDNA bearing the m.3243A>G, G allele mutation.
Reference:
- Kopinski, Piotr K., et al. "Mitochondrial DNA variation and cancer." Nature Reviews Cancer 21.7 (2021): 431-445.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Mitochondrial DNA variants can play inductive and adaptive roles in oncogenesis. (Kopinski et al., 2021)
Mitochondrial DNA variants can play inductive and adaptive roles in oncogenesis. (Kopinski et al., 2021) Human mtDNA map showing representative variants. (Kopinski et al., 2021)
Human mtDNA map showing representative variants. (Kopinski et al., 2021) Mitochondrial biology and metabolism and its relationship to the epigenome. (Kopinski et al., 2021)
Mitochondrial biology and metabolism and its relationship to the epigenome. (Kopinski et al., 2021)