Gene fragment analysis represents a cornerstone in contemporary genetic research, utilizing fluorescence-labeled DNA fragments in conjunction with capillary electrophoresis (CE) for their precise separation and detection. This sophisticated technique yields crucial information regarding DNA fragment size, relative quantification, and genotyping, thereby facilitating the identification of allelic variations, heterozygosity, chimerism, sample mixtures, and genetic relationships. Furthermore, gene fragment analysis is indispensable in applications such as microsatellite analysis and single nucleotide polymorphism (SNP) genotyping, providing vital insights into the understanding of genetic disorders.
The utility of gene fragment analysis extends to assessing the size and variability of defined genomic regions, encompassing insertions, deletions (indels), and SNPs. Such information is vital for unraveling the complex functions of genes and their regulatory mechanisms. For example, gene fragment analysis has been crucial in studies investigating the genes responsible for rice male reproductive development, where it has revealed gene expression patterns and regulatory networks. By meticulously analyzing specific genomic regions, researchers can deepen their understanding of gene function diversity and its impacts across various biological processes.
This article thoroughly explores the foundational principles and the wide-ranging applications of gene fragment analysis and detection technologies within genetic research. It examines essential methodologies such as DNA sequencing, polymerase chain reaction (PCR), and gel electrophoresis, highlighting their significant contributions across disciplines including oncology, neuroscience, infectious disease research, and agricultural science. Additionally, the article delves into the optimization of experimental designs, focusing on aspects such as PCR primer design and fluorescence labeling, aiming to enhance the precision and adaptability of gene fragment analysis.
Services you may interested in
Want to know more about the details of Gene Fragment Analysis? Check out these articles:
Basic Techniques for Gene Fragment Analysis
Gene fragment analysis employs a suite of core techniques including DNA sequencing, PCR, and gel electrophoresis combined with agarose gel analysis. The following sections provide a detailed examination of these methodologies:
A. DNA Sequencing
Sanger Sequencing
Sanger sequencing is a classical DNA sequencing technique based on the chain termination method, which utilizes dideoxynucleotide triphosphates (ddNTPs) to halt DNA synthesis, thereby producing DNA fragments of varying lengths. These fragments are separated and detected through capillary electrophoresis to generate sequence information. While Sanger sequencing may not match the throughput of Next-Generation Sequencing (NGS) technologies, it remains indispensable for small-scale sequencing projects, particularly in scenarios demanding high-quality sequence data.
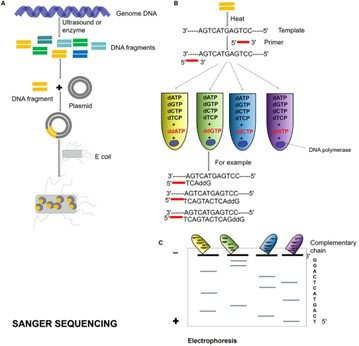 Sanger sequencing process. (Zhang, Lu, et al., 2021)
Sanger sequencing process. (Zhang, Lu, et al., 2021)
Next-Generation Sequencing (NGS)
NGS encompasses high-throughput sequencing technologies capable of concurrently sequencing millions of DNA fragments, markedly enhancing sequencing efficiency and resolution. This category includes platforms such as Illumina, PacBio, and Nanopore, which are suited for whole-genome sequencing, exome sequencing, and transcriptome analysis. The advantages of NGS lie in its high throughput, cost-effectiveness, and speed, though it also requires substantial bioinformatics support to manage and interpret the voluminous data it generates.
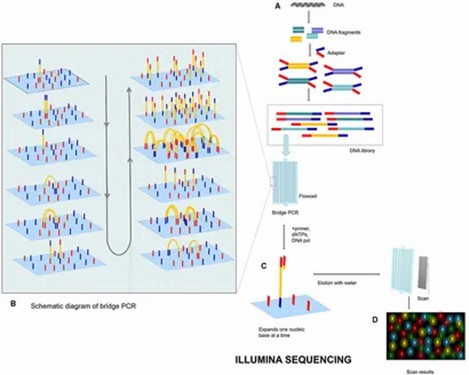 Illumina sequencing process. (Zhang, Lu, et al., 2021)
Illumina sequencing process. (Zhang, Lu, et al., 2021)
B. Polymerase Chain Reaction (PCR)
Conventional PCR
Conventional PCR is an analytical technique premised on DNA amplification, leveraging specific primer design to amplify target DNA fragments. This method finds extensive application in gene cloning, mutation detection, and gene expression analyses.
Quantitative Real-Time PCR (qPCR)
An advancement over conventional PCR, qPCR facilitates the real-time monitoring of fluorescence changes during amplification, allowing for the quantitative assessment of DNA or RNA copy numbers. qPCR is pivotal in gene expression analysis, pathogen detection, and studies of gene copy number variations.
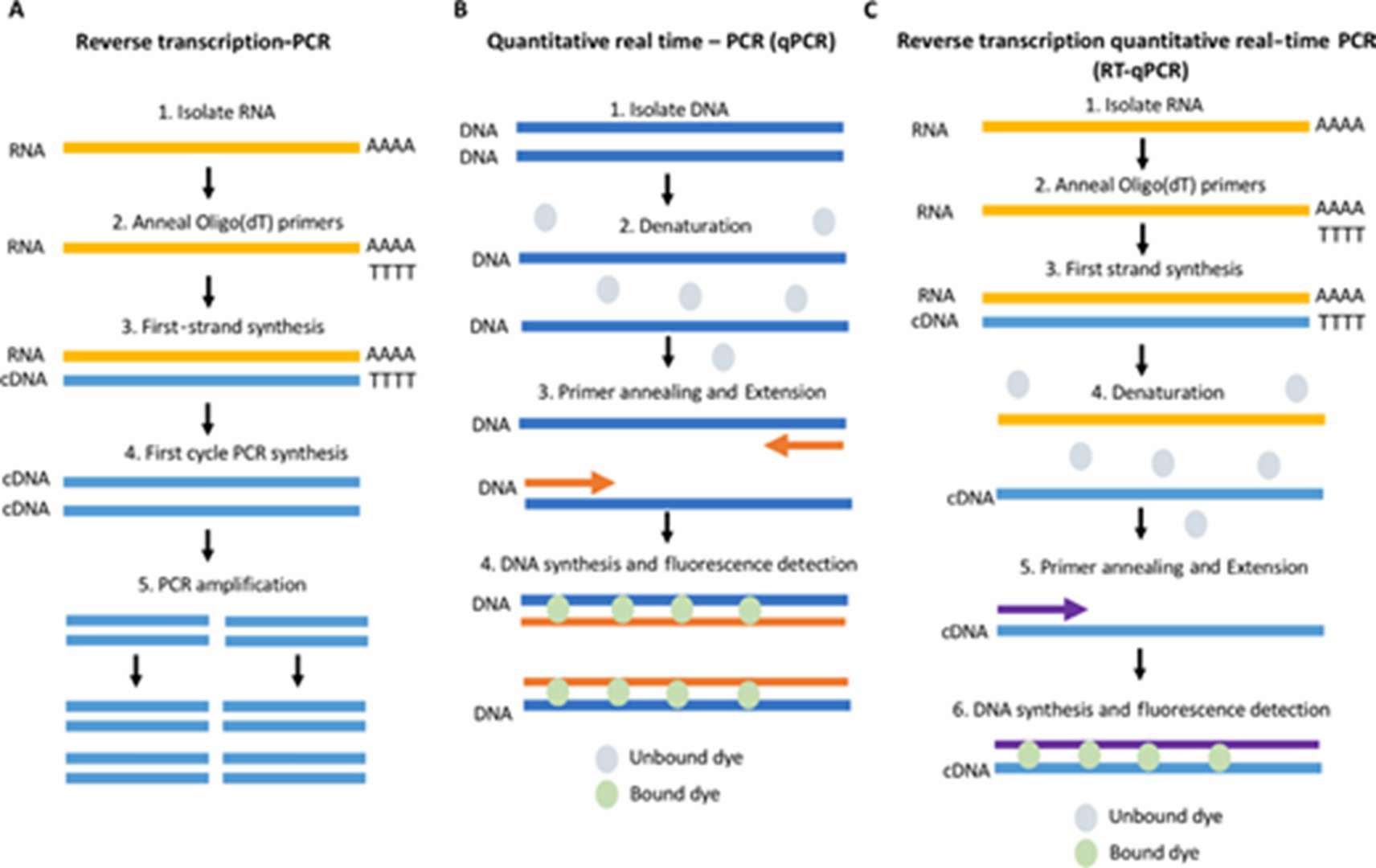 Schematic comparing RT-PCR, qPCR and RT-qPCR. (Adams, Grace, 2020)
Schematic comparing RT-PCR, qPCR and RT-qPCR. (Adams, Grace, 2020)
C. Gel Electrophoresis and Agarose Gel Analysis
Separation of Gene Fragments
Gel electrophoresis is a technique for separating DNA fragments based on molecular size, typically using agarose gel as the medium. Under an electric field, DNA fragments migrate through the gel, segregating according to size.
Visualization and Quantification
Post-separation, DNA fragments can be visualized using fluorescent dyes or radioactive labels, with fragment sizes determined via comparison with standard molecular weight markers. Gel imaging systems further facilitate quantitative analysis.
Conclusion
Each of these techniques possesses unique attributes:
- DNA Sequencing: Sanger sequencing is tailored for small-scale, high-quality sequencing, whereas NGS is geared towards large-scale, high-throughput sequencing.
- PCR: Conventional PCR is utilized for specific DNA fragment amplification, while qPCR is employed for quantitative analysis.
- Gel Electrophoresis: This method enables the separation and visualization of gene fragments and serves as a crucial ancillary tool in genetic analysis.
Collectively, these techniques form the bedrock of modern genomics research, providing robust tools for investigating gene functionality, diagnosing genetic disorders, and analyzing biodiversity.
Advanced Techniques for Gene Fragment Detection
Advanced gene fragment detection techniques span a range of methods, from bioinformatics tools to CRISPR systems, and include applications of mass spectrometry and proteomics. These technologies not only enhance the sensitivity and specificity of gene fragment detection but also offer powerful support for disease diagnosis, genetic research, and synthetic biology.
A. Bioinformatics Tools and Software
Sequence Alignment and Assembly
Bioinformatics tools are essential for genomic data analysis, including tasks such as genome comparison, genetic data analysis, and gene sequence alignment. Widely used software like BLAST and GenBank assists researchers in identifying variations and genetic differences within genomes. The advent of NGS technologies has markedly enhanced the efficiency of sequencing large-scale DNA fragments. Bioinformatics pipelines, such as BWA-MEM and GATK, are employed to process raw sequencing data, thereby improving analytical efficiency.
Annotation and Functional Prediction
Annotation tools facilitate the functional prediction of genome sequences, enabling tasks such as genome-wide mutation detection, visualization, and annotation using platforms like CRISPR-Detector. Additionally, CRISPR technologies are instrumental in evaluating gene editing efficiency, for instance, through the CRISPR-Cas9 system, which assesses editing outcomes for specific genes.
B. CRISPR-Based Detection Methods
CRISPR-Cas12 and CRISPR-Cas13 Systems
The CRISPR-Cas12 and CRISPR-Cas13 systems, recognized for their unique nucleic acid recognition capabilities, are widely used in gene fragment detection. Cas13a, for example, specifically cleaves RNA molecules and, when combined with isothermal amplification, enables highly sensitive nucleic acid detection. The Cas12 system facilitates the visualization of target DNA detection by cleaving DNA molecules and utilizing fluorescent reporters.
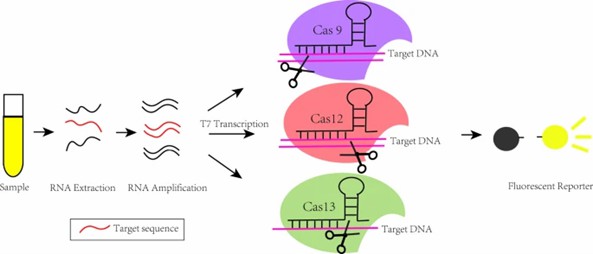 Based on the mechanism of CRISPR/Cas9, CRISPR/Cas12, CRISPR/Cas13 detection platform. (Lou, et al., 2022)
Based on the mechanism of CRISPR/Cas9, CRISPR/Cas12, CRISPR/Cas13 detection platform. (Lou, et al., 2022)
Applications in Gene Fragment Detection
CRISPR technology is employed across diverse applications in gene fragment detection, including pathogen detection, cancer research, and genetic disease diagnosis. Cas13a, coupled with isothermal amplification, allows for high-sensitivity RNA pathogen detection. Moreover, CRISPR-Cas9 is utilized in synthetic biology for cloning large DNA fragments, supporting research into biosynthetic gene clusters (BGCs).
C. Mass Spectrometry and Proteomics
Detection of Proteins Encoded by Gene Fragments
Proteomics techniques are used to detect proteins encoded by gene fragments. Mass spectrometry, for instance, is capable of identifying protein expression levels and integrates with bioinformatics tools for functional analysis.
Integration with Genomic Data
Proteomics data can be combined with genomic sequencing data to provide a comprehensive understanding of gene function and its roles in disease. By integrating CRISPR screening technologies with proteomics data, researchers can elucidate the impact of gene editing on cellular metabolism and signaling pathways.
Services you may interested in
Want to know more about the details of Gene Fragment? Check out these articles:
Workflow for Gene Fragment Synthesis and Analysis
The process of gene fragment synthesis and analysis is a multifaceted workflow encompassing multiple phases, beginning with sample preparation and culminating in data analysis. Each stage necessitates rigorous quality control and optimized protocols to ensure the validity and reliability of the resulting data. This systematic approach is indispensable not only for fundamental research but also for its substantial contributions to clinical diagnostics and advancements in biotechnology.
A. Sample Preparation
1. RNA Extraction and cDNA Synthesis
RNA extraction is the foundational step in sample preparation, typically involving the careful isolation of RNA from biological sources such as blood or tissue. This phase requires meticulous handling to maintain RNA integrity and purity, crucial for preventing contamination or degradation in downstream experiments. Once purified, the extracted RNA is reverse-transcribed into complementary DNA (cDNA) using reverse transcriptase. This process often includes selective mRNA amplification, which enhances RNA stability and improves the efficiency of subsequent amplification steps.
2. DNA Purification and Fragmentation
DNA purification and fragmentation are critical elements of the workflow. DNA can be fragmented through mechanical shearing methods, such as those employing Covaris instruments, or via enzymatic digestion, generating fragments appropriately sized for sequencing. Post-fragmentation, DNA undergoes end-repair and adapter ligation to aid in subsequent library construction.
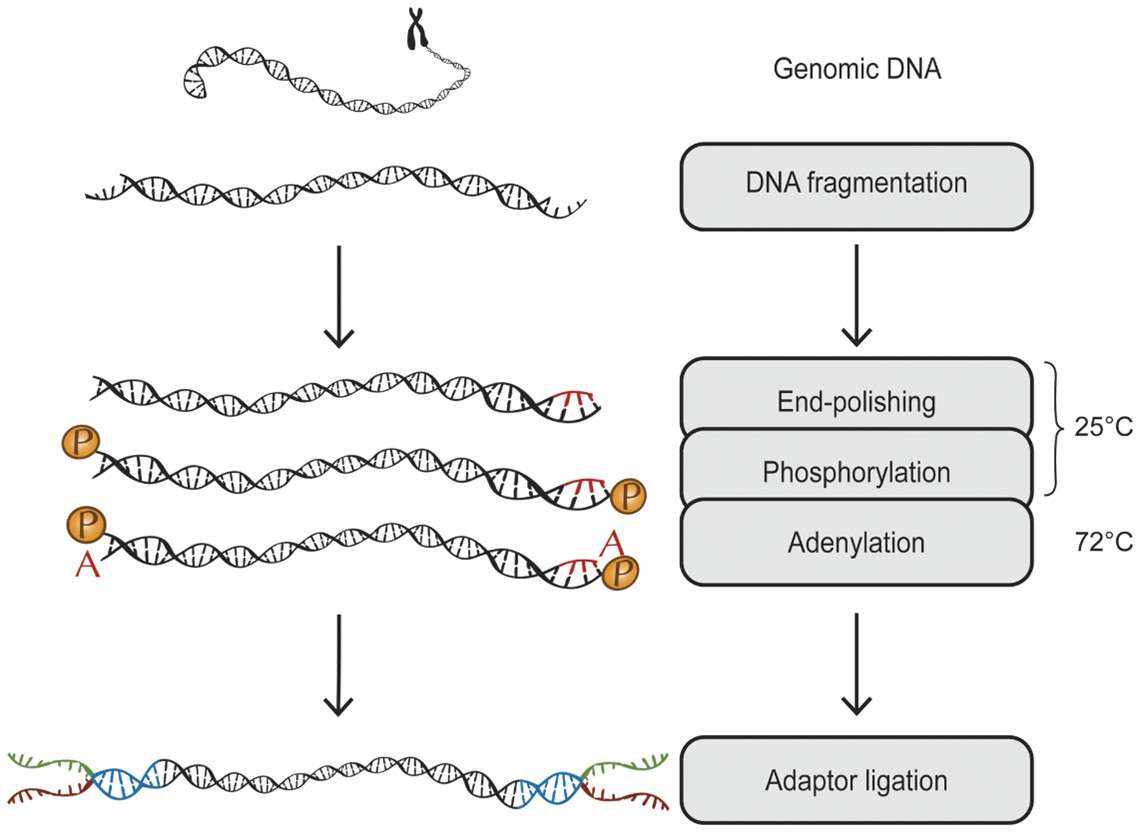 A schematic overview: genomic DNA is fragmentized, end-repaired, phosphorylated and adenylated in the same reaction. (Neiman, Mårten, et al., 2012)
A schematic overview: genomic DNA is fragmentized, end-repaired, phosphorylated and adenylated in the same reaction. (Neiman, Mårten, et al., 2012)
B. Library Construction and Sequencing
1. NGS Library Construction
Library construction transforms DNA or RNA samples into a format suitable for sequencing. This involves DNA fragmentation, end-repair, adapter ligation, and an assessment of library quality. Validation of library concentration and quality is performed using techniques like qPCR or electrophoresis, ensuring they meet the requirements of various sequencing platforms.
2. Sequencing Platforms and Protocols
The selection of a sequencing platform is contingent on specific research needs, with popular platforms including Illumina, Ion Torrent, PacBio, and Oxford Nanopore. Although sequencing protocols differ among platforms, they generally consist of raw data generation, quality control, and alignment to reference genomes.
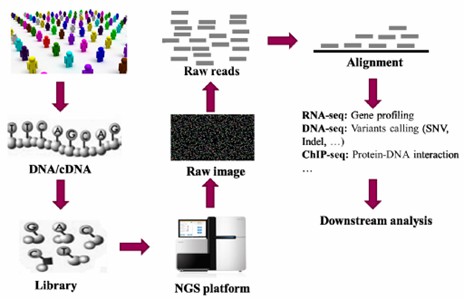 A brief flow chart of genetic studies using NGS. (Ye, Hao, et al., 2015)
A brief flow chart of genetic studies using NGS. (Ye, Hao, et al., 2015)
C. Data Analysis and Interpretation
1. Quality Control and Preprocessing
Following sequencing, the raw data must be subjected to stringent quality control to remove low-quality reads and adapter sequences. Commonly used tools for this purpose include FastQC and Trimmomatic. Preprocessing also involves trimming and standardizing reads to enhance the precision of subsequent analyses.
2. Identification of Gene Fragments and Isoforms
Aligning sequences to reference genomes or transcriptome databases facilitates the identification of gene fragments and their isoforms, such as splice variants. Tools like BWA and STAR are typically employed for alignment. Gene expression levels can be quantified using software like HTSeq or RSEM.
3. Functional and Quantitative Analysis
Upon identifying gene fragments and isoforms, further functional annotation and quantitative analysis are conducted. Gene Ontology (GO) analysis is employed to categorize gene functions, while differential expression analysis enables the comparison of gene expression across different samples. Additionally, techniques such as ChIP-seq and ATAC-seq can be integrated to explore gene regulatory mechanisms.
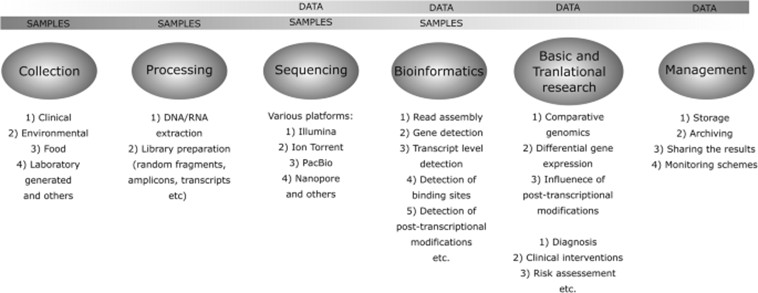 Overview of the different steps involved in the use of NGS technologies for data gathering and utilization. (Kozińska, et al., 2019)
Overview of the different steps involved in the use of NGS technologies for data gathering and utilization. (Kozińska, et al., 2019)
Case Studies of Gene Fragment Analysis
Gene fragment analysis techniques demonstrate substantial potential in model organism research and disease model analysis. Nonetheless, challenges such as the detection of low-abundance fragments and analysis of complex genomic regions persist. Through the optimization of PCR conditions, selective enrichment, and high-throughput sequencing, researchers can effectively overcome these obstacles, further advancing the field of genetic research.
A. Successful Applications of Gene Fragment Analysis Techniques
1. Identification of Novel Gene Fragments in Model Organisms
Gene fragment analysis techniques are extensively employed in studies involving model organisms such as mice and humans. By amplifying specific gene sequences via PCR and employing fluorescence labeling for separation, researchers can identify novel gene fragments. For instance, unique features within the TCRα and TCRβ loci have been discovered, enabling the identification of these gene fragments through recombination signal sequence (RSS) proximity, even in the absence of comprehensive gene annotations. Additionally, CRISPR-Cas9 technology has been leveraged in gene editing studies to precisely excise target DNA sequences, thereby advancing the understanding of gene functions in model organisms.
2. Functional Analysis of Gene Fragments in Disease Models
Gene fragment analysis techniques have yielded significant insights in disease model applications. For example, analyzing the fragment size distribution of circulating tumor DNA (ctDNA) enhances the sensitivity of tumor DNA detection. Furthermore, gene fragment analysis enables the detection of disease-associated mutations or changes in gene expression, providing crucial information for disease diagnosis and treatment strategies.
B. Challenges and Solutions in Gene Fragment Detection
1. Detection of Low-Abundance Gene Fragments
Detecting low-abundance gene fragments is a primary challenge in gene fragment analysis since the limited quantity of target fragments in samples is susceptible to background noise interference. To address this issue, researchers employ selective enrichment or computational analysis techniques to enhance detection efficiency. Techniques such as selective amplification or optimized PCR conditions significantly improve the sensitivity of detecting low-abundance fragments.
2. Analysis of Complex Genomic Regions
Another critical challenge is the analysis of complex genomic regions, which often contain repetitive sequences, highly homologous sequences, or areas that are difficult to amplify, leading to inaccuracies in gene fragment analysis results. Researchers have developed several strategies to tackle this problem, including the use of multiplex PCR, the design of specific primers, or the employment of high-throughput sequencing technologies to enhance the accuracy of analyzing complex regions.
Conclusion
Gene fragment analysis techniques-such as DNA sequencing, PCR, and gel electrophoresis-form the bedrock of modern genetic research. These methodologies are crucial for the identification of genetic variations, elucidation of gene functions, and examination of complex biological processes. The precise analysis of gene fragments is fundamental to advancing our understanding of genetic disorders, improving diagnostic techniques, and driving innovation in fields like agriculture and personalized medicine. Looking toward the future, continuous technological advancements promise to enhance the precision, speed, and accessibility of these techniques, thereby expanding their applications and deepening our understanding of genetics.
References:
- Al-Shuhaib, M.B.S., Hashim, H.O. Mastering DNA chromatogram analysis in Sanger sequencing for reliable clinical analysis. J Genet Eng Biotechnol 21, 115 (2023). https://doi.org/10.1186/s43141-023-00587-6
- Zhang, Lu, et al. "Advances in metagenomics and its application in environmental microorganisms." Frontiers in microbiology 12 (2021): 766364. https://doi.org/10.3389/fmicb.2021.766364
- Brockmöller, J., Tzvetkov, M.V. Pharmacogenetics: data, concepts and tools to improve drug discovery and drug treatment. Eur J Clin Pharmacol 64, 133–157 (2008). https://doi.org/10.1007/s00228-007-0424-z
- Adams, Grace. "A beginner's guide to RT-PCR, qPCR and RT-qPCR." The Biochemist 42.3 (2020): 48-53. https://doi.org/10.1042/BIO20200034
- Huang, Lei, et al. "CRISPR-detector: fast and accurate detection, visualization, and annotation of genome-wide mutations induced by genome editing events." Journal of Genetics and Genomics 50.8 (2023): 563-572. https://doi.org/10.1016/j.jgg.2023.03.010
- Lou, J., Wang, B., Li, J. et al. The CRISPR-Cas system as a tool for diagnosing and treating infectious diseases. Mol Biol Rep 49, 11301–11311 (2022). https://doi.org/10.1007/s11033-022-07752-z
- Neiman, Mårten, et al. "Library preparation and multiplex capture for massive parallel sequencing applications made efficient and easy." PLoS One 7.11 (2012): e48616. https://doi.org/10.1371/journal.pone.0048616
- Ye, Hao, et al. "Alignment of short reads: a crucial step for application of next-generation sequencing data in precision medicine." Pharmaceutics 7.4 (2015): 523-541. https://doi.org/10.3390/pharmaceutics7040523
- Kozińska, A., Seweryn, P. & Sitkiewicz, I. A crash course in sequencing for a microbiologist. J Appl Genetics 60, 103–111 (2019). https://doi.org/10.1007/s13353-019-00482-2
- Mouliere, Florent, et al. "Enhanced detection of circulating tumor DNA by fragment size analysis." Science translational medicine 10.466 (2018): eaat4921. DOI: 10.1126/scitranslmed.aat4921


 Sample Submission Guidelines
Sample Submission Guidelines
 Sanger sequencing process. (Zhang, Lu, et al., 2021)
Sanger sequencing process. (Zhang, Lu, et al., 2021) Illumina sequencing process. (Zhang, Lu, et al., 2021)
Illumina sequencing process. (Zhang, Lu, et al., 2021) Schematic comparing RT-PCR, qPCR and RT-qPCR. (Adams, Grace, 2020)
Schematic comparing RT-PCR, qPCR and RT-qPCR. (Adams, Grace, 2020) Based on the mechanism of CRISPR/Cas9, CRISPR/Cas12, CRISPR/Cas13 detection platform. (Lou, et al., 2022)
Based on the mechanism of CRISPR/Cas9, CRISPR/Cas12, CRISPR/Cas13 detection platform. (Lou, et al., 2022) A schematic overview: genomic DNA is fragmentized, end-repaired, phosphorylated and adenylated in the same reaction. (Neiman, Mårten, et al., 2012)
A schematic overview: genomic DNA is fragmentized, end-repaired, phosphorylated and adenylated in the same reaction. (Neiman, Mårten, et al., 2012) A brief flow chart of genetic studies using NGS. (Ye, Hao, et al., 2015)
A brief flow chart of genetic studies using NGS. (Ye, Hao, et al., 2015) Overview of the different steps involved in the use of NGS technologies for data gathering and utilization. (Kozińska, et al., 2019)
Overview of the different steps involved in the use of NGS technologies for data gathering and utilization. (Kozińska, et al., 2019)