DNA sequencing technologies have ushered in a new era within the biological sciences, helping us better understand into genomic architecture, evolutionary dynamics, pathogenic mechanisms, and beyond. As the genomics field burgeons, so too does the array of available sequencing technologies. This guide explains the main sequencing methods. It covers their basic principles, applications, and key factors to consider when choosing the right technology for your research.
1. What Are the Different Types of DNA Sequencing Technologies
DNA sequencing entails the precise determination of nucleotide sequences within a DNA molecule. Over recent decades, a plethora of sequencing techniques have emerged, each presenting distinct advantages and limitations. Choosing the right sequencing method depends on the investigative aims, sample characteristics, and the complexity inherent to the sequencing project.
Sanger Sequencing: The Cornerstone of DNA Analysis
Sanger sequencing, or chain-termination sequencing, remains the gold standard for low-throughput, targeted DNA sequencing endeavors. This method utilizes chain-terminating dideoxynucleotides (ddNTPs) to interrupt DNA strand elongation. Subsequent electrophoretic separation of resulting DNA fragments enables sequence determination based on termination points.
Despite its unparalleled accuracy and reliability for small-scale projects, Sanger sequencing's throughput remains comparatively limited. It is indispensable for sequencing smaller DNA regions, targeted assays, and validating findings generated by high-throughput sequencing technologies. Its superior capacity to resolve complex genome regions, particularly those harboring secondary structures, designates it as the preferred modality for detecting gene mutations and confirming variants.
Next-Generation Sequencing (NGS): Pioneering High-Throughput Genomic Exploration
NGS has changed genomic research. Unlike Sanger sequencing, which analyzes one DNA fragment at a time, NGS analyzes millions of fragments at once. These data culminate in the reconstruction of entire genomes or targeted regions of interest.
NGS is characterized by its high-throughput capabilities, enabling large-scale sequencing with expeditious turnaround times and cost-efficiency. Techniques including Illumina's sequencing-by-synthesis (SBS), Ion Torrent's semiconductor sequencing, and PacBio's Single Molecule Real-Time (SMRT) sequencing empower researchers to execute whole-genome sequencing (WGS) and targeted sequencing with unparalleled efficacy.
Long-Read Sequencing: Navigating Complex Genomic Landscapes
Long-read sequencing platforms, such as those developed by Oxford Nanopore and PacBio, offer an alternative to traditional short-read NGS technologies. These platforms can read substantially longer DNA fragments, spanning kilobases (kb) to megabases (Mb). This capability facilitates resolution of intricate genomic regions, including repetitive sequences, large structural variants, and genomic rearrangements that challenge short-read methodologies.
A notable advantage of long-read sequencing is its ability to yield contiguous assemblies with minimal gaps. This is invaluable for de novo genome assembly, structural variant detection, and exploration of complex genomic regions like telomeres and centromeres. However, long-read sequencing is often associated with higher error rates compared to short-read technologies, though ongoing advancements are progressively mitigating this discrepancy.
Single-Molecule Sequencing: Unraveling Individual DNA Strands
Single-molecule sequencing technologies, including PacBio's SMRT and Oxford Nanopore's nanopore sequencing, are leading the way. These approaches permit direct sequencing of individual DNA molecules sans amplification or fragmentation. In nanopore sequencing, DNA strands traverse a nanopore, with sequence determination based on alterations in electrical conductivity induced by nucleotides.
Single-cell sequencing offers clear benefits, such as directly sequencing long DNA fragments in real time. This makes it especially useful for detecting structural variations and other complex genomic features. Presently, single-molecule sequencing faces challenges related to higher error rates; however, continuous improvements in hardware and computational tools are enhancing its precision.
CD Genomics offers advanced high-throughput and long-read sequencing services, providing detailed insights into DNA and genome analysis for diverse research applications.
2. Sanger Sequencing: The Classic Approach
For over forty years, Sanger sequencing has served as the foundational technique in DNA sequencing, maintaining its status as a highly precise and dependable method, despite the emergence of more advanced technologies.
Mechanism of Sanger Sequencing
Sanger sequencing employs chain-terminating nucleotides, specifically dideoxynucleotide triphosphates (ddNTPs), which are devoid of the 3'-OH group required for DNA strand elongation. Within a reaction mixture comprising standard deoxynucleotide triphosphates (dNTPs) alongside limited amounts of each ddNTP, these terminators halt DNA synthesis at random points along the template strand. The resultant fragments are subsequently sorted by capillary electrophoresis, with sequence determination based on fragment lengths and the specific ddNTPs incorporated.
Applications of Sanger Sequencing
Sanger sequencing is ideally suited for applications necessitating high accuracy with low throughput, including:
- Targeted Sequencing: Sequencing discrete genes or specific genomic regions of interest.
- Mutation Detection: Identification of known genetic variants in clinical diagnostic settings.
- Validation of NGS Results: Confirmation of variants or findings that have been identified through high-throughput sequencing technologies.
Despite its precision, Sanger sequencing's limited throughput renders it less suitable for expansive projects, which demand extensive data generation. Nonetheless, it remains indispensable for precise, targeted applications such as single nucleotide polymorphism (SNP) analysis and validation of gene expression data.
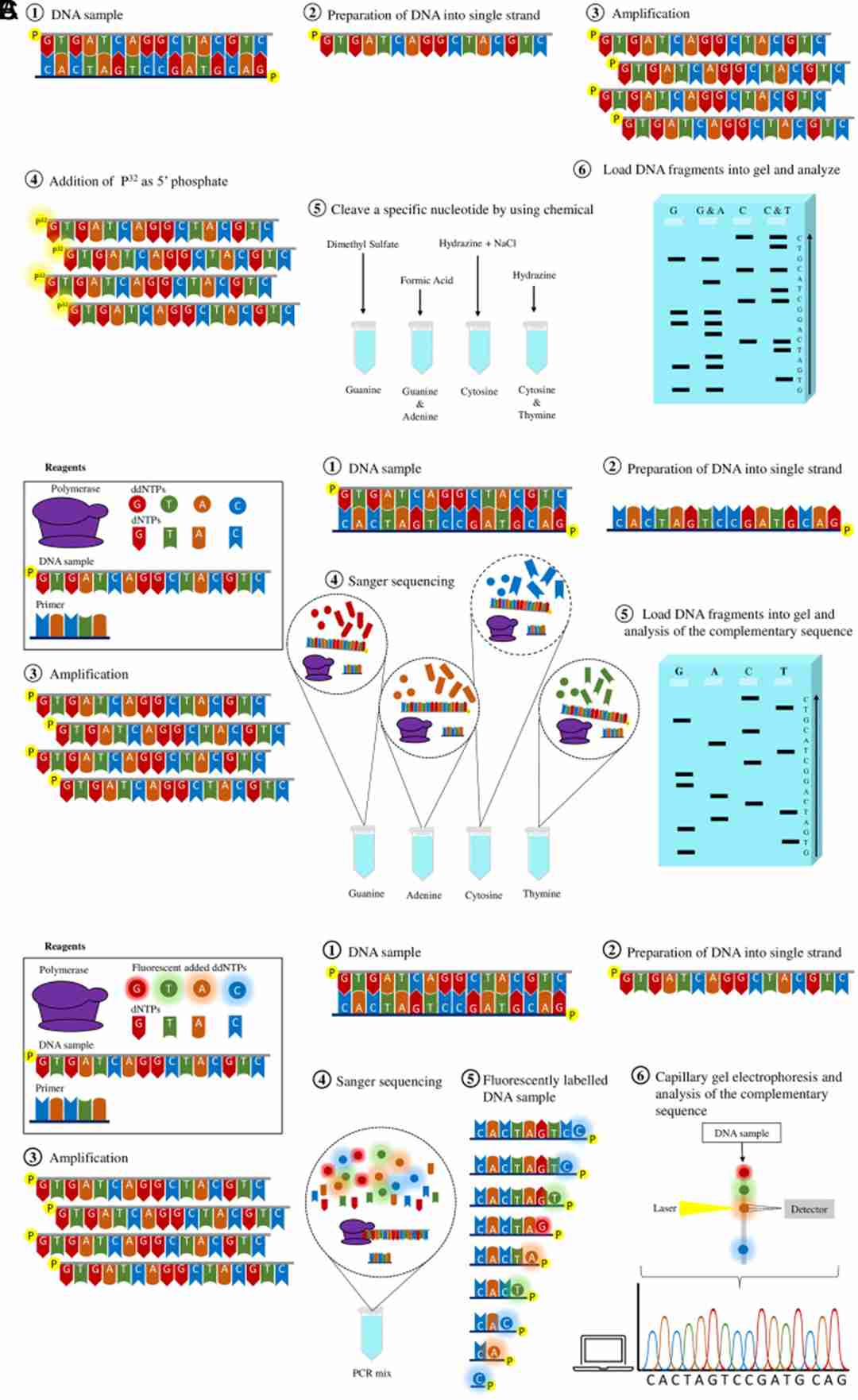 First-generation sequencing methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
First-generation sequencing methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
3. NGS: Revolutionizing Genomics
NGS technologies have remarkably redefined the breadth and depth of genomic analysis. Facilitating the concurrent sequencing of millions of DNA fragments, NGS significantly amplifies throughput while concurrently reducing costs, thus revolutionizing both research and clinical contexts.
Types of NGS Platforms
Several NGS platforms are extensively utilized, each conferring unique advantages contingent upon the specific sequencing objectives:
- Illumina (SBS technology): As the most prevalently employed platform, Illumina utilizes sequencing by synthesis (SBS), whereby nucleotides are appended to an expanding DNA strand and detected via fluorescent signals. Its high accuracy and diverse applicability, including WGS, RNA sequencing (RNA-Seq), and targeted sequencing, make it the preferred choice for large-scale sequencing undertakings.
- Ion Torrent: Employing semiconductor technology, Ion Torrent captures pH fluctuations resulting from nucleotide incorporation. It offers expedited sequencing speeds and is ideally suited for smaller-scale, cost-effective sequencing projects such as targeted sequencing and mutational analysis.
- PacBio (SMRT sequencing): PacBio's SMRT sequencing affords sequencing of extended DNA reads in real-time. This platform is instrumental for intricate genome assembly, structural variation analysis, and sequencing regions that pose challenges to short-read technologies.
- Oxford Nanopore: Utilizing nanopore technology, this platform facilitates direct DNA sequencing. As DNA traverses a nanopore, electrical current alterations are recorded, allowing nucleotide identification. Its real-time sequencing capability and proficiency in long-read sequences render it exceptionally versatile.
Applications of NGS
NGS is employed across a vast array of applications, including:
- WGS: Comprehensive genomic analysis across diverse organisms, encompassing human, microbial, or plant genomes.
- Whole-Exome Sequencing (WES): Targeted sequencing of genomic coding regions, critical for elucidating genetic diseases.
- RNA-Seq: Insight into gene expression profiles and identification of alternative splicing phenomena.
- Metagenomics: Analysis of microbial communities through NGS facilitates the exploration of biodiversity, microbiomes, and environmental genomics.
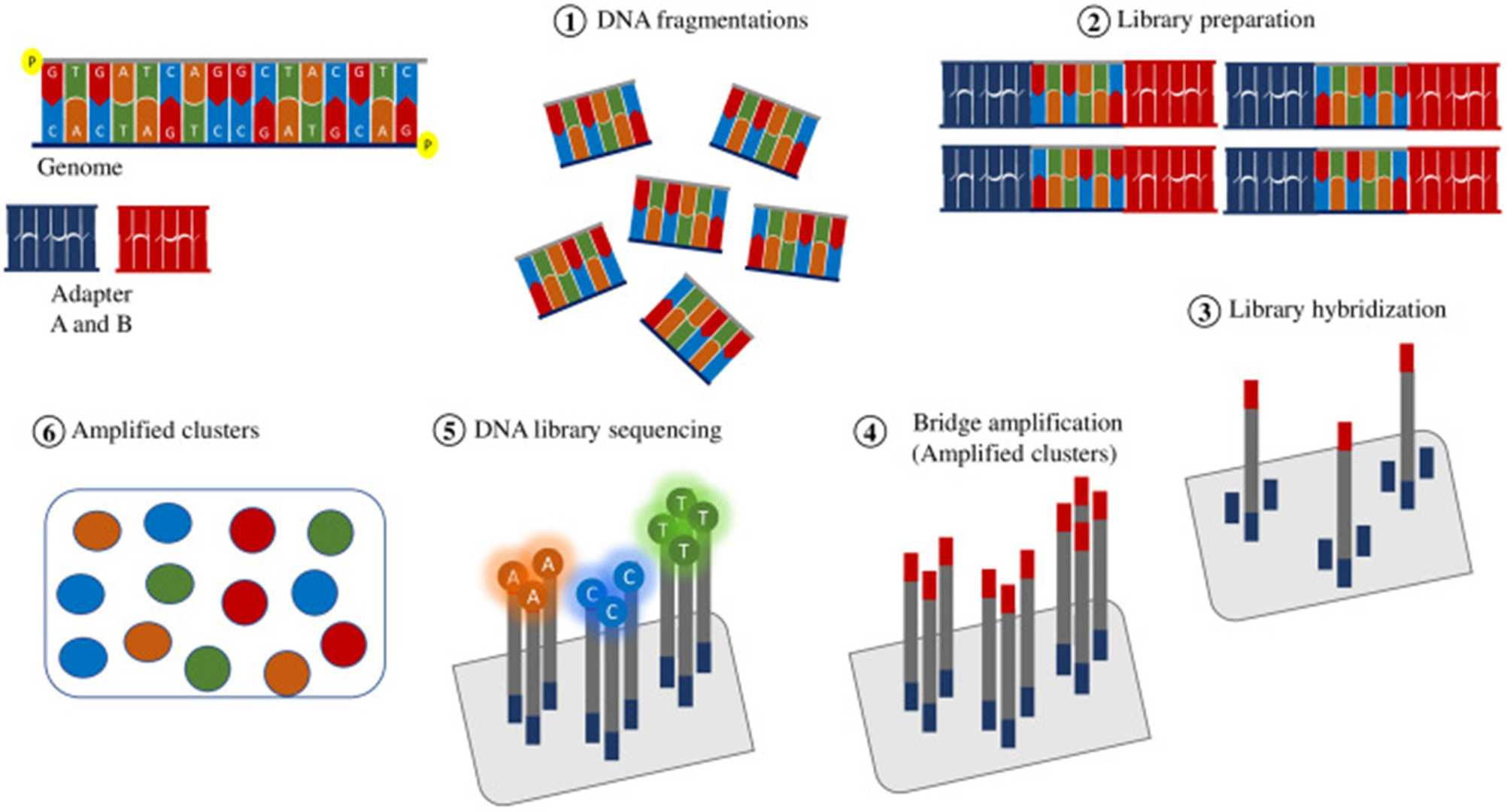 Library preparation and Illumina method. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
Library preparation and Illumina method. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
4. Beyond NGS: Emerging Technologies
In addition to classical sequencing methodologies, emerging technologies are broadening the horizons of DNA sequencing.
Long-Read Sequencing Technologies
Oxford Nanopore Technologies and PacBio have pioneered advancements in long-read sequencing, offering notable advantages over traditional short-read NGS. These advanced methods adeptly resolve complex genomic regions, including structural variants, repetitive sequences, and telomeres. The ability to generate real-time long reads makes these technologies invaluable for de novo genome assembly, human genome analyses, and extensive projects.
Single-Molecule Sequencing
Single-molecule sequencing platforms, such as PacBio's SMRT and Oxford Nanopore's nanopores, provide direct and real-time sequencing of extended DNA fragments. These platforms exhibit substantial promise for clinical genomics applications, where high accuracy and long reads are necessary for the elucidation of complex genetic disorders and structural variations.
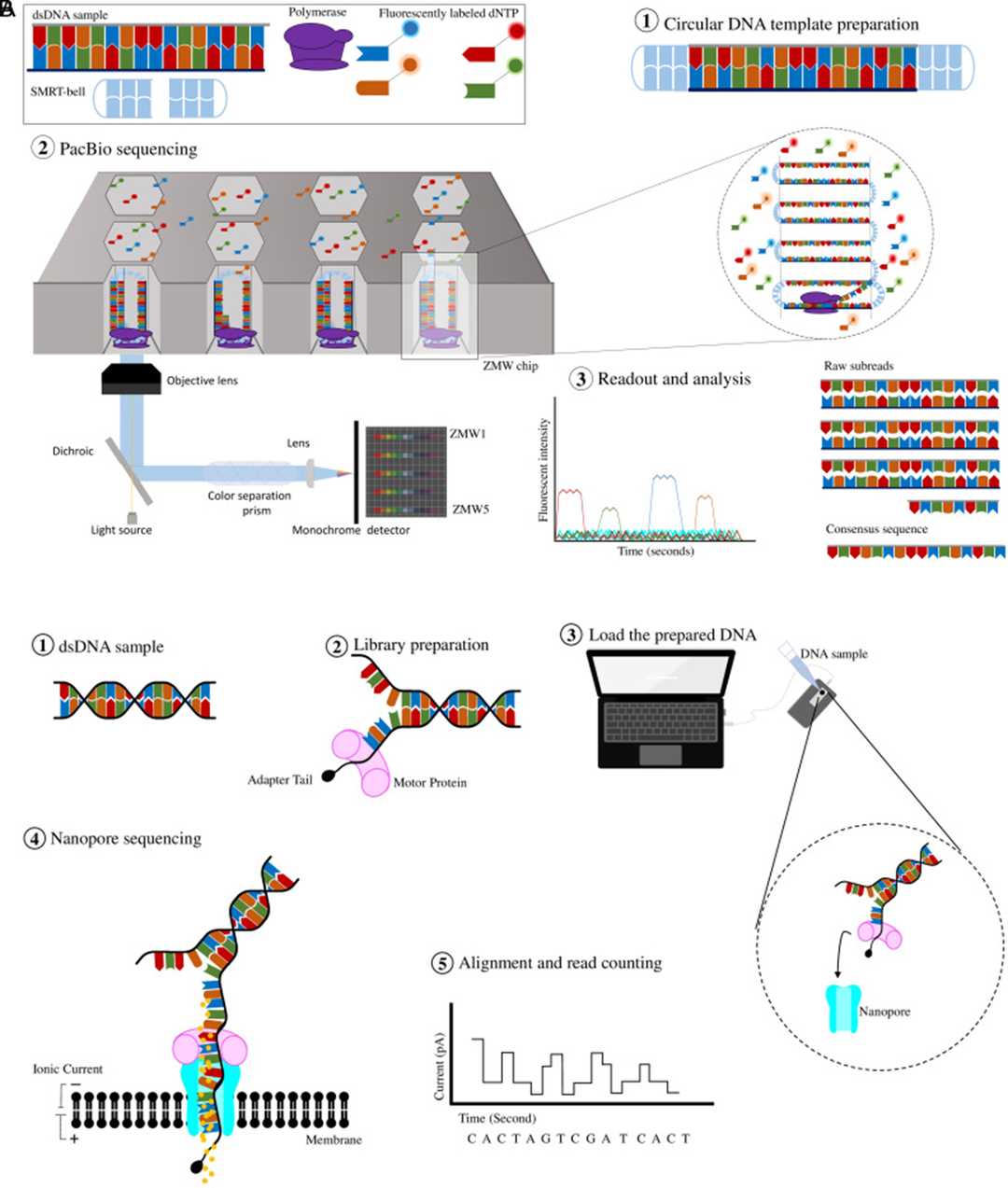 Third Generation Sequencing Methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
Third Generation Sequencing Methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
5. Common Sequencing Platforms and Their Devices
Each sequencing platform possesses unique strengths and limitations, rendering them as complementary instruments in modern genomics research. As sequencing technology improves, factors like cost, accuracy, and speed will keep influencing how each method is used.
Illumina Sequencing Platform
As a frontrunner in NGS, Illumina commands a significant market share globally, exceeding 70%. Utilizing sequencing-by-synthesis (SBS) technology, Illumina platforms deliver high throughput and precise sequencing across a wide spectrum of applications, including WGS, RNA-Seq, and targeted sequencing. Devices such as the MiSeq and MiniSeq are suited to small to medium-scale projects, exemplified by microbial genome sequencing and miRNA analysis, while the HiSeq and NovaSeq series are optimal for high-throughput endeavors like extensive genome studies.
Ion Torrent Platform
Pioneered by the developer of Roche 454 sequencing, the Ion Torrent platform introduced a groundbreaking semiconductor-based sequencing approach. By obviating the need for costly optical systems, Ion Torrent offers a more economical and expedited sequencing solution. The Ion Torrent PGM caters to small-scale projects like microbial or exome sequencing, whereas models like the Ion Proton and Ion S5 aim to enhance throughput capacity and manage larger datasets. Despite its operational efficiency and user-friendliness, the platform excels in targeted sequencing and clinical diagnostics owing to its comparatively lower throughput than Illumina.
MGI (BGI Genomics) Sequencing Platform
MGI, a division of BGI Genomics, has emerged as a formidable challenger in the sequencing domain. Leveraging technology acquired from Complete Genomics, MGI has developed sophisticated sequencing instruments like the MGISEQ-2000 and DNBSEQ-T7, concentrating on delivering high-throughput sequencing at reduced costs. These platforms are broadly employed in applications ranging from transcriptome sequencing to population-level investigations. MGI's market entry signifies an important shift towards diversifying the Illumina-dominated landscape, notably in regions such as China.
PacBio Sequencing (Pacific Biosciences)
Utilizing SMRT sequencing, PacBio platforms such as the Sequel II, generate long-read sequences with elevated accuracy. They are particularly fitting for de novo genome assembly, structural variant elucidation, and detailed characterization of complex genomic regions. While high per-sample costs persist, PacBio's capacity to deliver highly accurate long reads renders it indispensable for initiatives necessitating comprehensive genomic insights.
Oxford Nanopore Sequencing
Through the utilization of nanotechnology, Oxford Nanopore Technologies offers real-time sequencing solutions. Tools like the MinION are portable and versatile, proving suitable for fieldwork, rapid pathogen detection, and long genomic sequence analysis. Beyond DNA sequencing, Oxford Nanopore platforms can detect base modifications and structural variations, providing a distinct advantage for real-time and adaptive analysis. However, due to its lower accuracy relative to Illumina and PacBio, it is more suitable for niche applications such as epigenetics and structural genomics.
CD Genomics utilizes advanced sequencing platforms to deliver comprehensive insights across various research fields.
6. How to Choose the Right DNA Sequencing Method
Choosing an appropriate sequencing technology hinges on several factors including study scale, budgetary considerations, data quality demands, and specific research goals.
- For Small-Scale Projects: Sanger sequencing remains ideal for targeted sequencing and variant validation, offering high precision despite low throughput.
- For Large-Scale Studies: NGS technologies, especially Illumina and Ion Torrent, are optimal for high-throughput research such as WGS or RNA-Seq, delivering an equilibrium of cost-effectiveness, throughput, and accuracy.
- For Complex Genomes or Structural Variants: Technologies like PacBio and Oxford Nanopore, well-suited for challenging genomic regions and structural variant studies, should be considered.
Comparison of Sequencing Methods:
| Sequencing Method |
Strengths |
Limitations |
Best For |
| Sanger Sequencing |
High accuracy, simplicity, ideal for small projects |
Low throughput, unsuitable for large genomes |
Targeted sequencing, small genomic regions, verifying NGS results |
| NGS |
High throughput, cost-effective for large projects |
Higher upfront costs, complex workflows |
Whole-genome sequencing, RNA-seq, exome sequencing, variant detection |
| Long-Read Sequencing |
Resolves complex regions, detects large variants |
Higher error rates, expensive |
Complex genomes, structural variation, de novo assembly |
Ultimately, the decision depends on the research objectives, budget constraints, and required sequencing depth. In many contemporary research pursuits, combining methodologies—such as NGS for primary discovery followed by Sanger sequencing for validation—yields optimal results.
In conclusion, the field of DNA sequencing is continually evolving. Selecting the right sequencing method necessitates meticulous contemplation of project-specific needs. As a forerunner in genomics, CD Genomics offers an extensive range of sequencing services, leveraging state-of-the-art technologies to facilitate informed decision-making for researchers, ensuring the acquisition of high-quality, precise results.
References:
- Eren, Kübra, Nursema Taktakoğlu, and Ibrahim Pirim. "DNA sequencing methods: from past to present." The Eurasian journal of medicine 54.1 (2022): S47. doi: 10.5152/eurasianjmed.2022.22280
- Slatko, Barton E., Andrew F. Gardner, and Frederick M. Ausubel. "Overview of next‐generation sequencing technologies." Current protocols in molecular biology 122.1 (2018): e59. https://doi.org/10.1002/cpmb.59
- Van Dijk, Erwin L., et al. "The third revolution in sequencing technology." Trends in Genetics 34.9 (2018): 666-681. DOI: 10.1016/j.tig.2018.05.008
- Hu, Taishan, et al. "Next-generation sequencing technologies: An overview." Human Immunology 82.11 (2021): 801-811. https://doi.org/10.1016/j.humimm.2021.02.012


 Sample Submission Guidelines
Sample Submission Guidelines
 First-generation sequencing methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
First-generation sequencing methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022) Library preparation and Illumina method. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
Library preparation and Illumina method. (Kübra Eren et al., The Eurasian journal of medicine, 2022) Third Generation Sequencing Methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)
Third Generation Sequencing Methods. (Kübra Eren et al., The Eurasian journal of medicine, 2022)