The field of metagenomics enables researchers to investigate entire populations of microorganisms directly within their native habitats, circumventing traditional cultivation requirements. This revolutionary approach has catalyzed significant advancements across multiple scientific disciplines, from medical research and biological engineering to ecological studies and therapeutic development. Our detailed examination explores the fundamental principles underlying metagenomic analysis, including methodological approaches, practical implementations, and its transformative impact on contemporary scientific investigation.
Introduction to Metagenomics
What is Metagenomics?
Through direct analysis of environmental samples, metagenomics enables scientists to investigate complete microbial community genomes. Unlike conventional genomic approaches that examine single organisms' DNA sequences, this methodology encompasses entire populations. The collective genetic analysis reveals both taxonomic composition and metabolic capabilities of microorganisms within specific ecosystems.
This revolutionary technique has fundamentally altered our understanding of microbial ecosystems in different environments. Scientists can now decode the combined genomic content to reveal previously unidentified species and their ecological roles, surpassing the limitations of classical methods. The applications of metagenomic research have broad societal impact, from enhancing crop yields and developing novel therapeutics to implementing evidence-based conservation practices and advancing environmental protection measures.
Service you may interested in
Resource
Metagenomics in Understanding Microbial Diversity and Ecosystems
Through advanced DNA sequencing technologies, researchers can now investigate previously undetectable microorganisms that conventional cultivation methods miss. Metagenomic analysis of diverse environments - from terrestrial and aquatic systems to atmospheric samples and human-associated microbiota - illuminates the intricate relationships within microbial communities and their influence on ecological systems. These investigations yield valuable data about species diversity, pathogenesis, and environmental dynamics.
The field has transformed our approach to studying microbial ecology by facilitating direct examination of organisms within their native habitats. This methodology has enhanced our knowledge of how microorganisms impact both human wellness and environmental health.
Metagenomic studies have revealed previously unknown pathogens, identified novel antibiotic resistance genes, and contributed to our understanding of complex ecological processes such as methane and nitrogen cycling.
Metagenomics vs Genomics: Key Differences
Traditional Genomics vs. Shotgun Metagenomics
Genomics is centered on sequencing the genome of an individual organism, in contrast to metagenomics, which entails sequencing the DNA from a complete microbial community. A major difference between these two fields lies in the fact that genomics necessitates the isolation of a specific organism, while metagenomics can simultaneously gather genetic data from thousands of species without the need for culturing.
| Feature |
Traditional Genomics |
Shotgun Metagenomics |
| Target Organism |
Single species or individual genome |
Entire microbial community genome |
| Sample Requirement |
Requires isolation and culturing of the target species |
No culturing needed, DNA extracted directly from environmental samples |
| Applications |
Used to study the genetic structure and function of a single species |
Studies complex microbial communities, diversity, and ecological functions |
| Data Complexity |
Relatively simple, involves the genome of a single species |
Complex data with information from multiple microbial genomes |
| Technical Requirements |
Relatively low, traditional sequencing technologies suffice |
High-throughput sequencing and robust computational power required |
| Analysis Difficulty |
Lower, focused on analyzing a single species genome |
Higher, requires taxonomic classification and functional annotation of multiple species |
| Advantages |
Suitable for in-depth study of a specific species genome |
Can simultaneously study many microorganisms, ideal for environmental samples |
| Limitations |
Unable to effectively study complex microbial communities or environmental samples |
Faces challenges in data processing and analysis, particularly with genome assembly |
Why Metagenomics is the Preferred Choice for Studying Microbial Communities
Metagenomics has emerged as a preferred choice for studying microbial communities due to its ability to provide insights into environments where traditional culturing methods are not feasible. This approach is particularly valuable in exploring complex ecosystems such as the human gut, ocean microbiomes, and soil health. Metagenomics offers several advantages over traditional genomics, particularly in the study of microbial communities. Here are the key benefits:
Comprehensive Microbial Diversity Analysis:Metagenomics provides a broad view of microbial diversity by analyzing all genetic material in environmental samples, identifying unculturable microorganisms. Unlike traditional genomics, which focuses on culturing specific organisms, metagenomics reveals previously undetected species in environments like the human gut and ocean microbiomes.
Functional Insights into Microbial Communities:Metagenomics reveals not only the organisms present but also their functional roles by analyzing genes. This helps understand microbial metabolic pathways and ecological functions, such as identifying antibiotic resistance genes in clinical samples for treatment strategies.
Enhanced Resolution at Species and Strain Levels:Metagenomics offers higher resolution in taxonomy, identifying species and strains more accurately than traditional methods like 16S rRNA sequencing, which may miss closely related species. It can assemble complete genomes from mixed communities, providing precise microbial identification.
Reduced Bias from PCR Amplification:Unlike traditional genomic methods, which can introduce PCR amplification bias, metagenomics analyzes all DNA without amplification, offering a more accurate microbial community representation. This is crucial for accurate pathogen detection in clinical diagnostics.
Rapid and Cost-Effective Pathogen Detection:Metagenomic sequencing is faster and more sensitive than culture-based methods for pathogen detection, providing quicker diagnoses in clinical settings, which is critical for timely treatment and patient outcomes.
Ability to Detect Rare or Novel Pathogens:Metagenomics can identify rare or novel pathogens missed by traditional methods. It has successfully detected pathogens like in pneumonia cases where culturing failed, helping monitor emerging infectious diseases and public health threats.
Metagenomics Techniques: Tools for Understanding the Microbial World
Metagenomics employs various techniques to investigate microbial communities, with prominent methods including shotgun metagenomics, 16S rRNA sequencing, and metatranscriptomics. The suitability of each approach depends on the specific goals of the research, as each offers distinct advantages and constraints.
Key Metagenomic Techniques
Shotgun Metagenomics
Shotgun metagenomics, facilitates thorough examination of microbial ecosystems by simultaneously capturing genomic information from all present organisms. The resulting data yields insights into community composition, enzymatic capabilities, and biochemical processes operating within these populations. By analyzing complete genomic profiles, scientists can decode both taxonomic diversity and functional potential of complex microbial communities.
16S rRNA Sequencing
16S rRNA sequencing focuses on the 16S ribosomal RNA gene, which is present in all bacteria. It is commonly used for taxonomic profiling, enabling researchers to identify bacterial species present in a sample.See more in the 16S rRNA Sequencing vs. Metagenome Sequencing: A Guide for Selection.
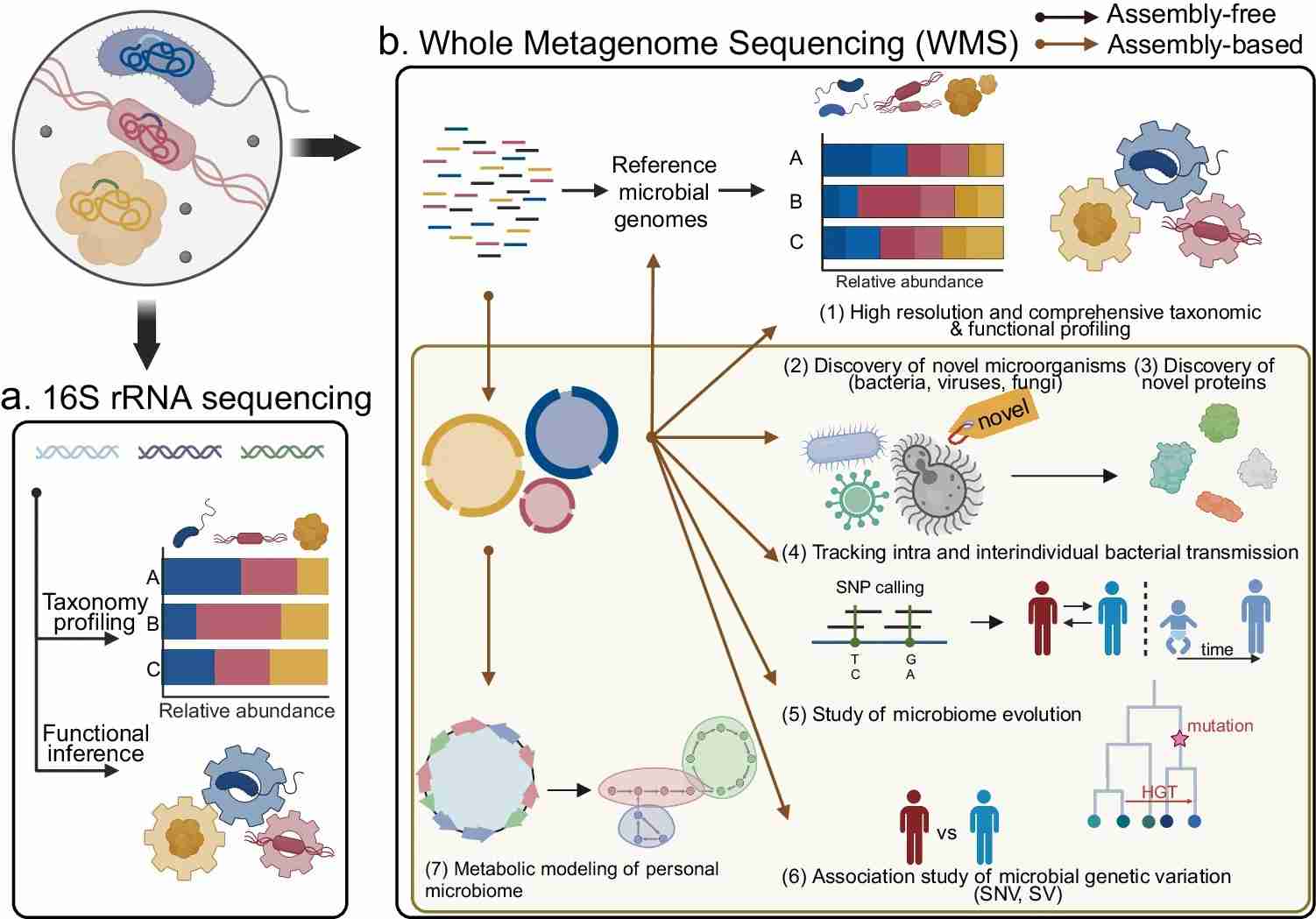 Fig. 1.Comparison of 16S rRNA sequencing and whole-metagenome sequencing (WMS) in microbiome analysis.(Quince, et.al. 2017)
Fig. 1.Comparison of 16S rRNA sequencing and whole-metagenome sequencing (WMS) in microbiome analysis.(Quince, et.al. 2017)
Metatranscriptomics
Metatranscriptomics focuses on the RNA transcripts present in a microbial community, providing insights into the gene expression and active metabolic pathways within that community. This technique helps understand the functionality of microbes in different environments.
Advantages and Limitations of Each Technique
Each metagenomic technique comes with its own set of benefits and challenges. Shotgun metagenomics provides the most comprehensive data but requires substantial computational resources. 16S rRNA sequencing is cost-effective and easier to analyze but offers limited functional insights. Metatranscriptomics provides detailed functional information but is more complex to analyze.
The Metagenomics Analysis Pipeline: From Data to Discovery
Metagenomics analysis involves a series of bioinformatics steps to process and interpret raw sequencing data. These steps typically include data preprocessing, taxonomic classification, and functional annotation.
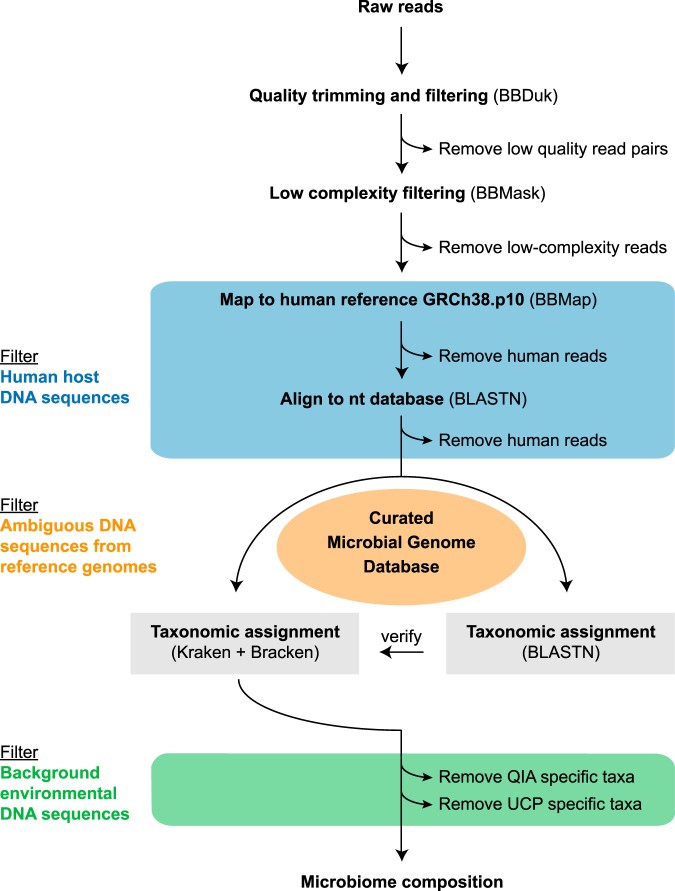 Fig. 2.Workflow for metagenomic data analysis.(Kirstahler, et.al. 2018)
Fig. 2.Workflow for metagenomic data analysis.(Kirstahler, et.al. 2018)
Key Stages of Metagenomics Analysis
Data Preprocessing
This step involves quality control and filtering to ensure that the sequencing data is reliable. This phase includes quality control measures such as removing low-quality sequences, adapter sequences, and host DNA contamination. For example, in a study analyzing gut microbiomes, researchers may use tools like FastQC to assess the quality of raw sequencing reads. If a significant number of reads are below the quality threshold (e.g., Q20), they may choose to trim these reads using Trimmomatic to enhance the overall dataset quality(Ibrahimi, et.al. 2023).
Taxonomic Classification
Taxonomic classification assigns sequences to specific taxa, helping identify microbial species or groups in a sample. Tools like MetaPhlAn enable researchers to classify microbial communities at the species level by comparing sequences to a comprehensive database of known microbial taxa. This approach has been successfully applied in environmental microbiome studies, uncovering previously uncharacterized species in diverse ecosystems(Blanco-Míguez,et.al. 2023).
Functional Annotation
By associating genetic sequences with their biological roles, researchers can decode the metabolic potential within microbial populations. Such analysis reveals how microorganisms participate in essential ecological processes, ranging from element transformation to defensive mechanisms and biosynthesis of active molecules. The identification of enzymatic systems, biochemical pathways, and genetic control mechanisms illuminates the fundamental drivers of microbial activities in ecosystems. Understanding these molecular mechanisms proves essential for harnessing microbial capabilities across diverse applications, from industrial biotechnology to therapeutic development and environmental remediation strategies.
Common Tools and Software
Several bioinformatics tools are integral to metagenomics analysis. Tools such as QIIME facilitate taxonomic classification through 16S rRNA gene sequencing, while Kraken offers rapid taxonomic classification based on k-mer analysis. For functional annotation, HUMAnN and MetaPhlAn are widely used to analyze metabolic pathways and community composition respectively.
Applications of Metagenomics
The implementation of metagenomic techniques spans multiple sectors, transforming industrial processes through novel biofuel development, identification of catalytic proteins, and production of bioactive compounds. Within medical research, this methodology enhances our capacity for clinical diagnostics, analysis of host-associated microbiota, and surveillance of emerging pathogens and antimicrobial resistance patterns. Environmental applications encompass monitoring ecological conditions and anthropogenic effects, while agricultural utilization focuses on enhancing soil microbiome management, promoting plant resilience, and developing biological control strategies. The breadth of these practical applications demonstrates how metagenomic analysis has become fundamental across numerous scientific and industrial domains.
 Fig. 3.Contributions of metagenomics to various fields. (Nam, et.al. 2023)
Fig. 3.Contributions of metagenomics to various fields. (Nam, et.al. 2023)
Biotechnology and Industry
Metagenomics is extensively applied in biotechnology, particularly in areas such as biofuel production, enzyme discovery, and the synthesis of natural products. For instance, researchers have utilized metagenomic techniques to identify novel enzymes from uncultured microorganisms in extreme environments, which are capable of catalyzing reactions under harsh conditions. An example is the discovery of extremozymes from deep-sea sediments that can function at high temperatures and pressures, making them valuable for industrial processes that require robust catalysts.
Healthcare and Medical Research
In the healthcare sector, metagenomics is transforming the landscape of disease diagnosis, microbiome research, and pathogen identification. A notable application is in the Human Microbiome Project (HMP), which has provided insights into the complex microbial communities residing in various human body sites. This project has revealed associations between specific microbiome profiles and health conditions, such as obesity and inflammatory bowel disease. Additionally, metagenomics is crucial for detecting antibiotic resistance genes in clinical samples, enabling healthcare providers to tailor treatments more effectively.
Environmental Science
Metagenomics plays a pivotal role in analyzing environmental microbiomes across diverse ecosystems, including soil, water, and air. For example, metagenomic studies of marine sediments have led to the discovery of previously unknown antibiotic-producing bacteria that could be harnessed for new pharmaceuticals.Furthermore, by assessing microbial diversity and functional potential in various habitats, researchers can monitor ecosystem health and evaluate the impacts of human activities.
Agriculture
In agriculture, metagenomics is employed to enhance soil health, manage crop disease resistance, and optimize pest control strategies. By investigating soil microbiomes, researchers have identified beneficial microorganisms that promote plant growth and suppress diseases.
Case Study of mapping the metagenomic diversity in glacier-fed stream
Background
This study investigates glacier-fed streams (GFS), one of Earth's most extreme aquatic ecosystems, characterized by oligotrophy and environmental fluctuations. Microorganisms primarily exist in biofilm forms, but how they adapt to these conditions remains unclear. Utilizing 156 metagenomes from sediment samples of GFS across nine mountain ranges from the "Vanishing Glaciers" project, the study uncovered thousands of metagenome-assembled genomes (MAGs), including prokaryotes, algae, fungi, and viruses.
Results
The GFS environments were characterized by extreme oligotrophic conditions, with low nutrient concentrations and minimal algae biomass. Metagenomic sequencing of sediment samples uncovered a wealth of microbial life, including 2,868 high-quality bacterial MAGs, with 21% of genera previously unreported. The findings also revealed methane-producing archaea, suggesting GFS as a potential source of methane. Overall, this research highlights the unique microbial ecosystems of glacier-fed streams and provides new insights into microbial adaptations to harsh environments.
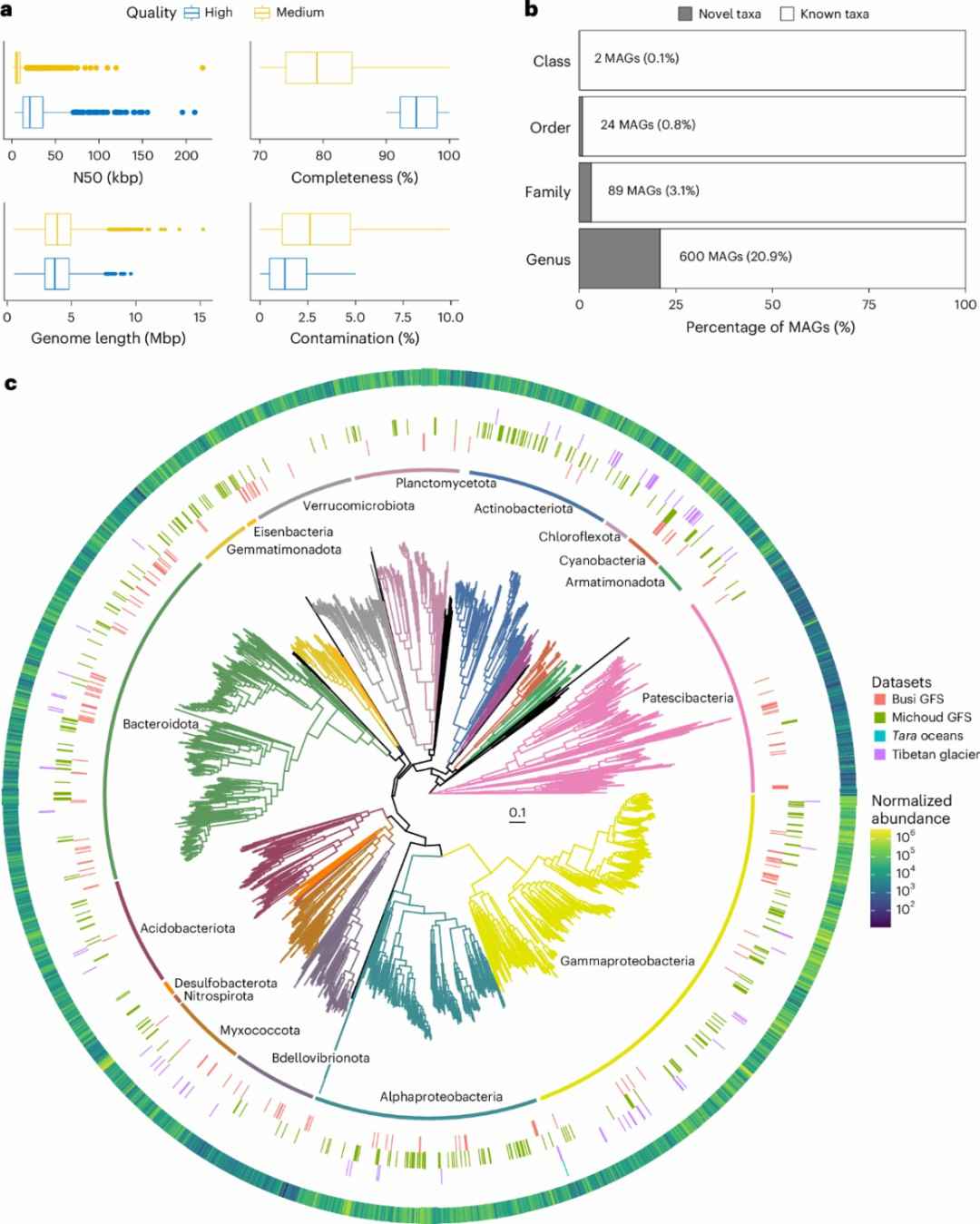 Fig. 4.MAGs of Prokaryotes from glacier-fed stream. (Michoud, et.al. 2025)
Fig. 4.MAGs of Prokaryotes from glacier-fed stream. (Michoud, et.al. 2025)
Conclusion
The emergence of metagenomic analysis has fundamentally transformed our understanding of microbial populations, creating unprecedented opportunities across scientific fields. Ongoing developments in DNA sequencing platforms, coupled with enhanced computational capabilities, suggest even greater potential for future discoveries. This powerful methodology continues to decode complex microbial interactions within natural systems, advancing numerous domains from therapeutic development and biological engineering to ecological conservation and beyond.
References:
- Quince, C., Walker, A., Simpson, J. et al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 35, 833–844 (2017). https://doi.org/10.1038/nbt.3935
- Kirstahler, P., Bjerrum, S. S., et.al. (2018). Genomics-Based Identification of Microorganisms in Human Ocular Body Fluid. Scientific reports, 8(1), 4126. https://doi.org/10.1038/s41598-018-22416-4
- Ibrahimi, E., Lopes, M. B.,et.al. (2023). Overview of data preprocessing for machine learning applications in human microbiome research. Frontiers in microbiology, 14, 1250909. https://doi.org/10.3389/fmicb.2023.1250909
- Blanco-Míguez, A., Beghini, F., Cumbo, F. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat Biotechnol 41, 1633–1644 (2023). https://doi.org/10.1038/s41587-023-01688-w
- Nam, N. N., Do, H. D. K.,et.al. (2023). Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions. Foods (Basel, Switzerland), 12(11), 2140. https://doi.org/10.3390/foods12112140
- Michoud, G., Peter, H., Busi, S.B. et al. Mapping the metagenomic diversity of the multi-kingdom glacier-fed stream microbiome. Nat Microbiol 10, 217–230 (2025). https://doi.org/10.1038/s41564-024-01874-9


 Sample Submission Guidelines
Sample Submission Guidelines
 Fig. 1.Comparison of 16S rRNA sequencing and whole-metagenome sequencing (WMS) in microbiome analysis.(Quince, et.al. 2017)
Fig. 1.Comparison of 16S rRNA sequencing and whole-metagenome sequencing (WMS) in microbiome analysis.(Quince, et.al. 2017) Fig. 2.Workflow for metagenomic data analysis.(Kirstahler, et.al. 2018)
Fig. 2.Workflow for metagenomic data analysis.(Kirstahler, et.al. 2018) Fig. 3.Contributions of metagenomics to various fields. (Nam, et.al. 2023)
Fig. 3.Contributions of metagenomics to various fields. (Nam, et.al. 2023) Fig. 4.MAGs of Prokaryotes from glacier-fed stream. (Michoud, et.al. 2025)
Fig. 4.MAGs of Prokaryotes from glacier-fed stream. (Michoud, et.al. 2025)