Overview of Bulk Segregant Analysis
Definition of Bulk Segregant Analysis (BSA)
Bulk Segregant Analysis, is a gene mapping approach rooted in the principles of population genetics. At its core, BSA involves constructing one or more pooled samples (referred to as "bulks") from individuals exhibiting extreme phenotypes. By comparing the genomic differences between these bulks, researchers can rapidly pinpoint molecular markers or genes closely linked to the target traits. BSA is extensively employed in the genetic mapping studies of both plants and animals, proving particularly valuable in the localization of major genes associated with quantitative and qualitative traits.
Basic Workflow of BSA:
- Parental Population Construction: Select individuals displaying extreme phenotypes as parents.
- Bulk Construction: Combine the DNA of individuals with extreme phenotypes to form multiple bulks.
- High-Throughput Sequencing: Sequence the bulk DNA to analyze SNP or InDel markers across the genome.
- Data Analysis: Identify trait-associated loci by calculating genotype frequency differences of SNP or InDel markers.
- Functional Annotation: Perform functional annotation of candidate genes to further explore their roles in trait regulation.
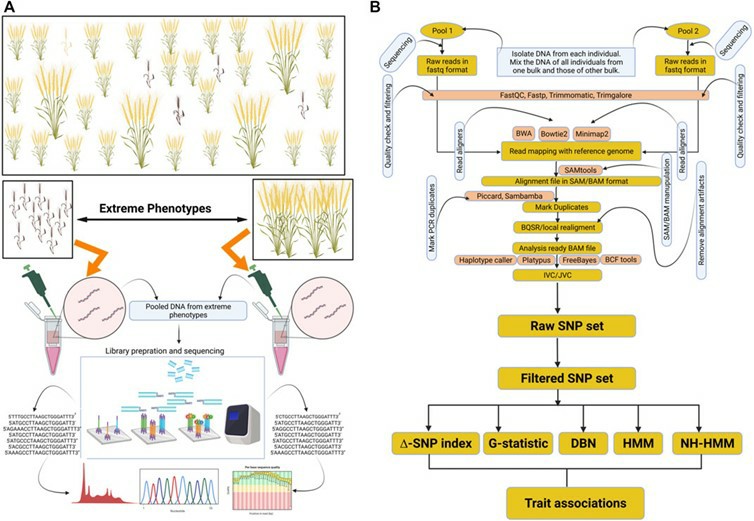 Representation of BSA-seq and general data analysis approach for marker trait associations. (Majeed, Aasim, et al. 2022)
Representation of BSA-seq and general data analysis approach for marker trait associations. (Majeed, Aasim, et al. 2022)
Historical Background and Development of BSA
Introduced by Michelmore and colleagues in 1991, BSA was conceived as a rapid and cost-effective gene mapping method aimed at quickly identifying genes or molecular markers related to specific traits by isolating individuals with extreme phenotypes from a population.
- Early Applications: Initially applied in plant genetics to map disease resistance genes in crops like wheat and Arabidopsis, BSA found utility in increasingly complex organisms, including humans and animals, as high-throughput sequencing technologies advanced.
- Technical Improvements: With the proliferation of next-generation sequencing (NGS), BSA coupled with high-throughput sequencing (BSA-seq) emerged as a mainstream approach. This integration not only increased the resolution of gene mapping but also reduced the experimental timeframe.
- Modern Applications: Recently, BSA has been combined with deep learning algorithms (e.g., DeepBSA) to further enhance analytical precision, enabling the mapping of genes associated with complex traits. BSA-seq is also utilized in crop breeding and genetic enhancement to identify key genes controlling significant agronomic traits.
- Future Prospects: As computational power and sequencing technologies continue to evolve, BSA is anticipated to achieve high-precision gene mapping across a broader range of species, thereby supporting precision agriculture and biomedical research.
Since its inception, BSA has evolved from traditional methods to the incorporation of high-throughput sequencing technologies, becoming a pivotal tool in the field of gene mapping. Its efficiency and cost-effectiveness have facilitated its widespread application in the fields of plant and animal genetics, genomics, and breeding research.
Services you may interested in
The Science Behind BSA
BSA serves as an efficient gene mapping technique by constructing bulks and utilizing high-throughput sequencing to swiftly identify molecular markers or gene regions linked to target traits. Compared to traditional methods, BSA's efficiency, cost-effectiveness, and flexibility make it especially suitable for resource-limited contexts. However, its lower resolution and reliance on extreme phenotypes present limitations. The future integration of NGS and deep learning algorithms is anticipated to enhance the precision and applicability of BSA, broadening its scope of use.
1. Gene Mapping Methods Based on BSA
Bulk Segregant Analysis is a rapid, efficient, and cost-effective technique for gene mapping that primarily targets the identification of molecular markers or QTLs associated with specific phenotypes. The fundamental principle of BSA involves selecting individuals with extreme phenotypes from segregating progenies of parental lines to construct two distinct bulks, each representing a genotype combination related to the extreme phenotypes. Subsequently, high-throughput sequencing technologies, such as RNA-Seq or whole-genome resequencing, are employed to sequence these bulks. By comparing genomic differences between them, researchers can identify molecular markers or gene regions associated with the trait of interest.
Specific Steps Involved:
- Bulk Construction: From segregating populations, individuals with extreme phenotypes are selected to construct two bulks, R and S, each comprising individuals with similar phenotypes.
- Sequencing and Analysis: The R and S bulks undergo high-throughput sequencing. By comparing their genomic variations, polymorphic sites (e.g., SNPs or Indels) significantly associated with phenotypic differences are screened.
- Localization and Validation: The identified molecular markers undergo association analysis with the target trait, which narrows down the candidate regions. The results are further validated using marker-assisted selection (MAS) or traditional linkage analysis.
The strength of BSA lies in its efficiency and cost-effectiveness, particularly in scenarios with small population sizes or limited experimental resources.
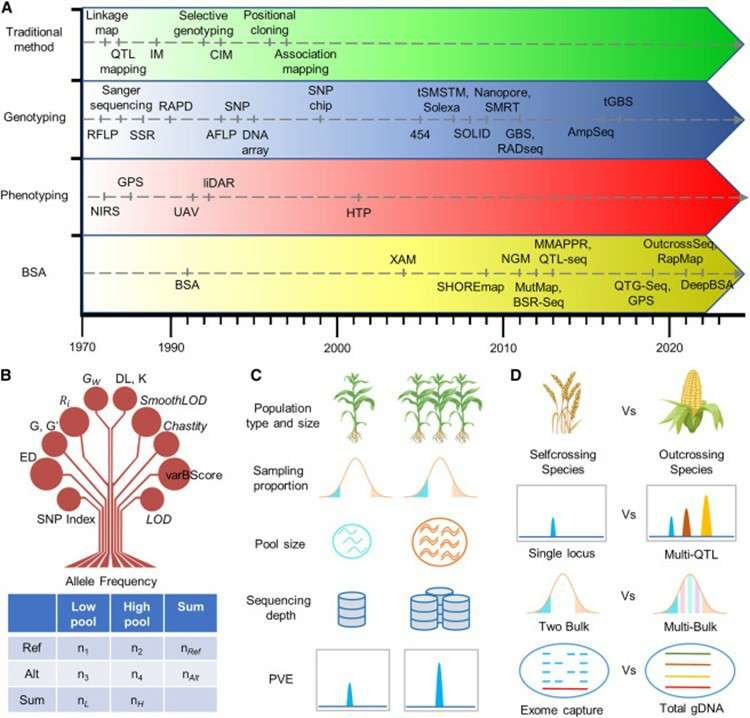 Comprehensive overview of BSA. (Wang, Xi, et al., 2023).
Comprehensive overview of BSA. (Wang, Xi, et al., 2023).
2. Comparison of BSA with Traditional Gene Mapping Methods
- Traditional Gene Mapping Methods: These typically depend on the creation of large segregating populations (such as F2 populations) and utilize linkage analysis to develop genetic maps and pinpoint target genes. Despite their precision, these methods are time-consuming and costly, requiring extensive samples and intricate experimental designs.
- Advantages of BSA:
- Efficiency: BSA requires only a small number of individuals with extreme phenotypes to construct bulks, significantly reducing sample demands and experimental duration.
- Cost-effectiveness: With fewer individuals per bulk, sequencing costs are considerably reduced.
- Flexibility: BSA is applicable to any segregating population, including F2 populations of outcrossing species or parental lines of self-pollinating crops.
- Broad Applicability: BSA is effective for both qualitative traits (e.g., disease resistance) and quantitative traits (e.g., yield).
- Limitations:
- Lower Resolution: Limited numbers of individuals per bulk may lead to reduced mapping precision.
- Reliance on Extreme Phenotypes: The extreme phenotypes of individuals in the bulks might be influenced by environmental factors, affecting result accuracy.
- Selection Bias of Polymorphic Sites: There might be a bias towards certain regions, causing potentially significant areas to be overlooked.
Comparison of Gene Mapping Methods
| Method |
Applicability |
Cost-effectiveness |
Technical Platform |
| Bulked Segregant Analysis |
Suitable for any segregating population, including biparental and outcrossing populations, particularly effective for rapidly detecting molecular markers associated with specific traits. |
Significantly reduces sequencing and analysis costs, simplifies the sequencing process. |
Utilizes next-generation sequencing (NGS), such as RNA-Seq and whole-genome re-sequencing. |
| Traditional QTL Mapping |
Suitable for large segregating populations requiring genetic linkage maps. |
High cost and time-consuming, requiring numerous samples and extensive marker screening. |
Employs traditional molecular marker technologies, such as RFLP and SSR. |
| QTL-seq |
Suitable for qualitative traits and quantitative traits with significant major genes. |
High cost, but offers high-resolution QTL mapping. |
Based on NGS technology. |
| MutMap |
Designed for mutant analysis. |
High cost, but provides high-resolution mutant mapping. |
Uses NGS technology. |
| MutMap+ |
Optimal for early-lethal or non-heterozygous mutants. |
High cost, offers high-resolution mutant mapping. |
Relies on NGS technology. |
| MutMap-gap |
Appropriate for traits where the target gene is not present in the reference genome. |
High cost, provides high-resolution QTL mapping. |
Utilizes NGS technology. |
| OcBSA |
Particularly suitable for QTL mapping in outcrossing populations. |
High cost, delivers high-resolution QTL mapping. |
Based on NGS technology. |
| DeepBSA |
Ideal for QTL mapping and functional gene cloning of complex traits. |
High cost, provides high-resolution QTL mapping. |
Incorporates deep learning algorithms. |
Designing and Implementing BSA Experiments
The design and execution of BSA experiments demand a rigorous approach, beginning with the formulation of research questions and extending through sample selection, data processing, and statistical analysis. Through meticulous experimental design and robust data analysis, the complex relationships between genes and phenotypes can be illuminated, providing crucial insights for subsequent research endeavors. Below is a detailed exposition:
1. Key Steps in Experimental Design
Grounded in a wealth of evidence, the design of an experiment typically comprises the following critical steps:
- Clarifying the Research Question: Establish the primary objectives and research questions of the experiment, such as investigating the relationship between genes and phenotypes.
- Defining Variables: Identify the independent variables (e.g., genotypes) and dependent variables (e.g., phenotypes), and elucidate their interrelationships.
- Formulating the Experimental Plan: Determine the logistics, including the timing, location, subjects, equipment, and instruments.
- Selecting Experimental Methods and Tools: Choose appropriate methods and tools based on the experiment's objective, such as randomization and control group setups.
- Planning for Data Collection and Analysis: Define data collection methods, processing techniques, and statistical analysis tools to be employed.
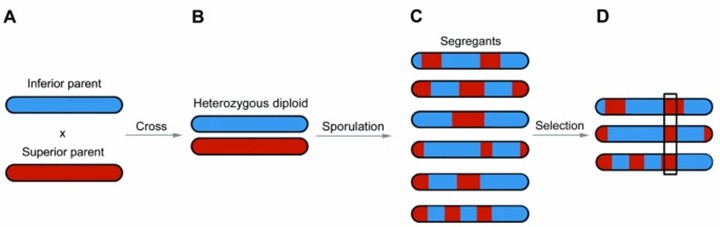 Bulk segregant analysis for mapping genomic regions linked to a phenotype of interest in yeast. (Duitama, J. et al., 2014)
Bulk segregant analysis for mapping genomic regions linked to a phenotype of interest in yeast. (Duitama, J. et al., 2014)
2. Sample Selection and Processing
In BSA experiments, careful selection and handling of samples are critical:
- Source of Samples: Samples are typically derived from natural populations or artificially generated segregating populations, such as F2 populations.
- Sample Processing: Conduct necessary preprocessing steps like DNA extraction, PCR amplification, or protein extraction to ensure data reliability and representativeness.
- Random Grouping: Assign samples randomly to different experimental groups to minimize bias and enhance reproducibility.
- Controlling Variables: Strictly control environmental conditions and other potential confounding variables throughout the experiment.
3. Data Analysis and Interpretation
Data analysis forms the core of BSA experiments:
- Data Preprocessing: Clean and organize the collected data by removing outliers and imputing missing values.
- Statistical Analysis: Employ suitable statistical techniques (e.g., chi-square tests, t-tests, or regression analysis) to examine data and verify hypotheses.
- Interpreting Results: Interpret the relationship between genotypes and phenotypes based on data analysis, proposing possible biological mechanisms.
- Visualization: Present the results of data analysis through graphical representations to facilitate an intuitive understanding of trends and relationships.
4. Iterative Nature of Experimental Design
BSA experimental design is inherently iterative and requires continual refinement based on observed results:
- Preliminary Testing: Conduct pilot tests before the main experiment to validate the effectiveness of the design.
- Feedback and Adjustment: Adjust the experimental setup, refine variable settings, and optimize data processing methods based on pilot test outcomes.
- Final Validation: Verify the reliability and consistency of results through repeated experimentation upon completion of all experimental phases.
5. Considerations in Experimental Design
When designing and executing BSA experiments, several considerations must be observed:
- Avoiding Bias: Ensure the absence of human-induced bias through random grouping and blind procedures.
- Indicator Validation: Verify that the chosen indicators accurately reflect the expected outcomes.
- Ethical Review: Obtain approval from an ethics review board when biological samples or living subjects are involved.
Want to know more about the details of BSA? Check out these articles:
Applications of BSA in Plant Research
BSA is a highly efficient technique for gene mapping, playing a critical role in plant genetics and crop improvement. When combined with other advanced technologies, BSA's scope of application is expected to broaden, offering a more robust toolset for plant research and breeding programs.
1. Applications of BSA in Plant Genetics
BSA is an effective gene mapping technique that swiftly identifies trait-associated gene loci by analyzing pools of individuals with extreme phenotypes. Its primary advantages include:
- Efficiency: Compared to traditional map-based cloning methods, BSA significantly reduces experimental costs and time.
- Applicability: The technique is suitable for polyploid plants, plants with complex genomes like cotton and wheat, and perennials.
- Flexibility: BSA can be combined with high-throughput sequencing technologies, such as BSA-seq, to enhance mapping precision.
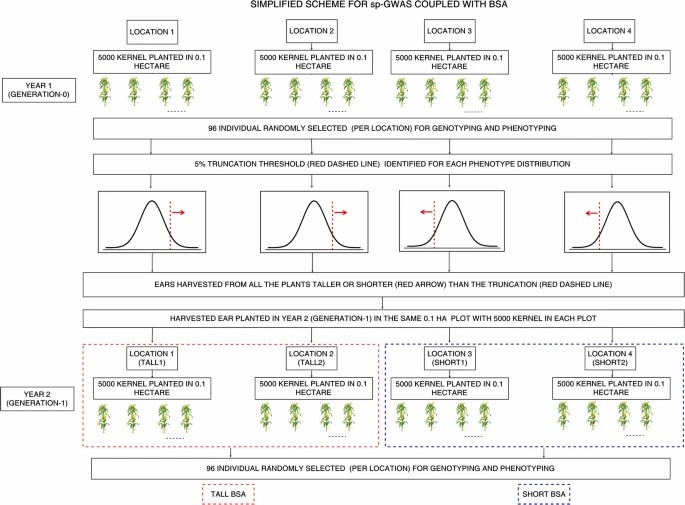 Schematic pipeline of sp-GWAS coupled with BSA. (Gyawali, A. et al., 2019)
Schematic pipeline of sp-GWAS coupled with BSA. (Gyawali, A. et al., 2019)
The application of BSA spans a wide range of agricultural traits, including disease resistance, stress tolerance, and yield attributes. For example, in maize, BSA has been used to identify the candidate gene OSARF1 associated with dwarfing; in rice, it successfully mapped the OsARF1 gene related to dwarf phenotypes.
2. Case Studies of BSA in Crop Improvement
- Disease Resistance Research: BSA is extensively utilized for mapping plant disease resistance genes. In 1991, Michelmore et al. used BSA to successfully identify the DM5/8 gene associated with late blight resistance in potatoes. Similarly, in citrus fruits, BSA, combined with molecular markers, has been used to map disease resistance genes.
- Yield Trait Studies: BSA demonstrates remarkable efficacy in mapping yield-related traits. In maize, BSA combined with genome-wide association studies (GWAS) has enabled the identification of candidate genes related to plant height. In soybeans, BSA has been used to map genes associated with chlorosis mutations.
- Stress Resistance Research: BSA also finds extensive application in stress resistance studies. In wheat, for instance, BSA, paired with molecular markers, has facilitated the identification of QTLs related to salt stress tolerance.
3. Advantages and Limitations of BSA in Gene Identification
Advantages:
- Rapid and Efficient: BSA allows for the swift analysis of a large number of samples, making it suitable for large-scale gene mapping studies.
- Cost-effective: Compared to traditional map-based cloning, BSA reduces the number of experimental steps and resource consumption.
- Broad Applicability: It is suitable for the localization of both single and multiple traits and is relatively unaffected by environmental factors.
Limitations:
- Dependence on Extreme Phenotypes: BSA requires selection of individuals with extreme phenotypes for pooled analysis, which may limit its use in complex traits.
- Inability to Detect Epistasis: BSA primarily targets major genes and may not capture complex gene interactions.
- High Sample Quality Requirements: The phenotype and genotype of samples need to be highly consistent; otherwise, result accuracy may be compromised.
4. Combining BSA with Other Technologies
To overcome its limitations, researchers have integrated BSA with other techniques:
- Integration with GWAS: Combining single-plant GWAS (sp-GWAS) with BSA enhances the accuracy of candidate gene identification.
- Incorporation of Transcriptomic Analysis: Pairing RNA-seq with BSA allows further exploration of gene functions.
- Inclusion of Deep Learning Algorithms: Recently, deep learning-enhanced BSA software (e.g., DeepBSA) has been developed to improve data analysis efficiency and accuracy.
5. Future Perspectives
With advancements in sequencing technology, BSA holds great potential for expansion. Future research directions include:
- Multi-omics Integration: Combining BSA with transcriptomics, proteomics, and other omics data for comprehensive gene function analysis.
- High-throughput Analysis: Utilizing NGS to further enhance the resolution and efficiency of BSA.
- Application Across Species: Expanding the use of BSA to non-model plants, such as woody and perennial species.
Such developments promise to extend the utility and effectiveness of BSA in plant genetics and beyond.
If you want to learn more about BSA applications, you can refer to the article "Applications of Bulk Segregant Analysis in Plant Research."
Conclusion
Bulk Segregant Analysis represents a highly efficient and cost-effective approach to gene mapping, enabling the rapid identification of genes associated with specific traits. Its straightforward methodology and resource efficiency render it invaluable across genetic studies involving a wide array of organisms. The integration of BSA with next-generation sequencing, known as BSA-seq, enhances the precision and broadens the applicability of this technique, thereby propelling advancements in fields such as plant breeding and medical research. As technological progress continues to unfold, BSA remains a pivotal tool in the realm of genetic discovery and innovation.
References:
- Edae, Erena A., and Matthew N. Rouse. "Bulked segregant analysis RNA-seq (BSR-Seq) validated a stem resistance locus in Aegilops umbellulata, a wild relative of wheat." PLoS One 14.9 (2019): e0215492. https://doi.org/10.1371/journal.pone.0215492
- Vendrely, Katelyn M., et al. "Humanized mice and the rebirth of malaria genetic crosses." Trends in parasitology 36.10 (2020): 850-863. DOI: 10.1016/j.pt.2020.07.009
- Majeed, Aasim, et al. "Harnessing the potential of bulk segregant analysis sequencing and its related approaches in crop breeding." Frontiers in Genetics 13 (2022): 944501. https://doi.org/10.3389/fgene.2022.944501
- Wambugu, Peterson, et al. "Sequencing of bulks of segregants allows dissection of genetic control of amylose content in rice." Plant Biotechnology Journal 16.1 (2018): 100-110. https://doi.org/10.1111/pbi.12752
- Wang, Xi, et al. "Next-generation bulked segregant analysis for Breeding 4.0." Cell Reports 42.9 (2023). https://doi.org/10.1016/j.celrep.2023.113039
- Song, Jian, et al. "Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean." Frontiers in plant science 8 (2017): 919. https://doi.org/10.3389/fpls.2017.00919
- Gyawali, A., Shrestha, V., Guill, K.E. et al. Single-plant GWAS coupled with bulk segregant analysis allows rapid identification and corroboration of plant-height candidate SNPs. BMC Plant Biol 19, 412 (2019). https://doi.org/10.1186/s12870-019-2000-y
- Duitama, J., Sánchez-Rodríguez, A., Goovaerts, A. et al. Improved linkage analysis of Quantitative Trait Loci using bulk segregants unveils a novel determinant of high ethanol tolerance in yeast. BMC Genomics 15, 207 (2014). https://doi.org/10.1186/1471-2164-15-207


 Sample Submission Guidelines
Sample Submission Guidelines
 Representation of BSA-seq and general data analysis approach for marker trait associations. (Majeed, Aasim, et al. 2022)
Representation of BSA-seq and general data analysis approach for marker trait associations. (Majeed, Aasim, et al. 2022) Comprehensive overview of BSA. (Wang, Xi, et al., 2023).
Comprehensive overview of BSA. (Wang, Xi, et al., 2023). Bulk segregant analysis for mapping genomic regions linked to a phenotype of interest in yeast. (Duitama, J. et al., 2014)
Bulk segregant analysis for mapping genomic regions linked to a phenotype of interest in yeast. (Duitama, J. et al., 2014) Schematic pipeline of sp-GWAS coupled with BSA. (Gyawali, A. et al., 2019)
Schematic pipeline of sp-GWAS coupled with BSA. (Gyawali, A. et al., 2019)