In the intricate domain of immunology, a profound understanding of the immune repertoire is pivotal for progressing in the development of therapeutics, vaccines, and diagnostic methodologies. At the heart of this field are two critical elements: B-cell receptors (BCRs) and T-cell receptors (TCRs). These receptors, while both indispensable for recognizing pathogens, engage in distinctly divergent mechanisms that profoundly affect their sequencing and subsequent analysis. BCRs are adept at recognizing intact, unprocessed antigens, whereas TCRs primarily engage with processed peptide fragments presented by major histocompatibility complex (MHC) molecules. This fundamental divergence not only influences their structural characteristics but also dictates their individual functional roles within the immune system. As we delve into the methodologies and challenges surrounding BCR and TCR sequencing, these differences emerge as crucial to deepening our comprehension of immune responses. A deeper appreciation of these distinctions is anticipated to ultimately enhance the development of more targeted and efficacious interventions for immune-related disorders, paving the way for personalized treatment strategies for patients.
BCR vs TCR Sequencing: Methodologies and Challenges
Sequencing the BCR and TCR repertoires offers valuable insights into immune responses. However, there are key differences in how these sequences are obtained and analyzed, as demonstrated by recent scientific studies. Before delving into the difference between TCR and BCR sequencing, it's essential to understand the structure and function of TCRs and BCRs. For more details, please refer to the article "Differences Between BCR and TCR ".
BCR Sequencing:
Methodology: The sequencing of BCR commonly employs multiplex PCR to amplify specific immunoglobulin genes, alongside 5' rapid amplification of cDNA ends (5' RACE) to extend the variable regions. In a landmark study, Briney et al. effectively combined multiplex PCR with high-throughput sequencing to analyze the human antibody repertoire, showcasing the efficacy of this approach in capturing the extensive diversity of BCRs.
Challenges: The process of BCR sequencing is inherently complex due to the significant diversity introduced by somatic hypermutation, necessitating a markedly greater sequencing depth to obtain reliable and comprehensive data. Wardemann and Busse have underscored the intricate challenges associated with BCR sequencing, notably in adequately capturing the breadth of somatic hypermutation and faithfully representing the antibody repertoire's diversity.
TCR Sequencing:
Methodology: The sequencing of T-cell receptors primarily employs PCR amplification techniques, with a critical focus on the alpha and beta chains of the receptor. Advanced methodologies, such as RNA sequencing (RNA-Seq), are frequently utilized for detailed TCR profiling. A pivotal study by Rosati et al. undertook a meticulous comparison of various TCR sequencing approaches, including targeted amplicon sequencing and RNA-Seq. This study elucidated the unique advantages inherent to each method in the context of TCR repertoire analysis.
Challenges: TCR sequencing is relatively less encumbered by issues of diversity than B-cell receptor (BCR) sequencing, generally necessitating a lower sequencing depth. Nevertheless, significant challenges persist, particularly in the precise quantification of clonal frequencies and the differentiation of closely related sequences, as highlighted by Heather et al.
Both BCR and TCR sequencing methodologies have undergone significant evolution to surmount specific technical hurdles. The integration of molecular barcodes represents a notable advancement, enhancing the fidelity of sequencing by mitigating PCR and sequencing errors. This innovation, as demonstrated by Khan et al., has considerably refined the accuracy of immune repertoire analyses.
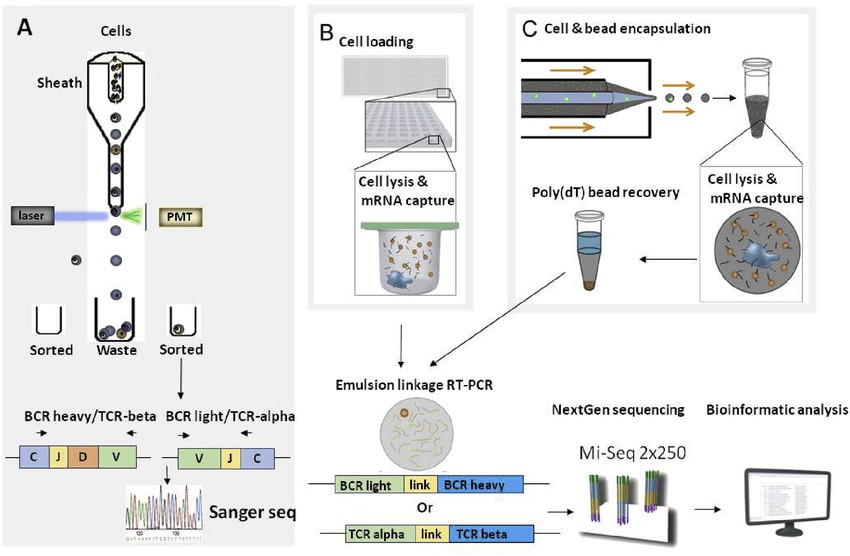 Figure 1: Approaches for BCR and TCR repertoire sequencing. (Yu Fen Samantha Seah et al,. 2017)
Figure 1: Approaches for BCR and TCR repertoire sequencing. (Yu Fen Samantha Seah et al,. 2017)
For more on the method of TCR profiling, visit the Strategy Overview for Next-Generation Sequencing based TCR Profiling. For details on immune receptor sequencing, check out our overview of immune repertoire sequencing.
Service you may intersted in
Comparison of TCR and BCR Sequencing Platform
| Sequencing Technology |
Accuracy (%) |
Read Length (bp) |
Cost (per million bases) |
Throughput (reads/run) |
| Next-Generation Sequencing (NGS) |
99.5 |
300 |
100 |
106−107106−107 |
| PacBio |
99.0 |
10,000 - 15,000 |
2000 |
Varies |
| Oxford Nanopore |
95.0 |
Up to 100,000 |
Varies |
Varies |
Applications: NGS is widely used for comprehensive BCR and TCR repertoire analysis due to its high throughput and lower cost per base. It allows for the analysis of large sample sizes, making it suitable for studies in cancer immunotherapy and autoimmune diseases.
Limitations: While NGS provides good accuracy, the read lengths are shorter compared to long-read technologies, which may complicate the assembly of full-length TCR and BCR sequences.
Applications: PacBio sequencing offers long read lengths that are advantageous for resolving complex regions of TCR and BCR genes. This technology is particularly useful for applications requiring full-length sequencing of variable regions in immune receptors.
Limitations: The higher cost associated with PacBio sequencing may limit its use in large-scale studies compared to NGS.
Applications: Oxford Nanopore technology enables real-time sequencing with exceptionally long reads, which can capture entire TCR and BCR sequences in a single read. This capability is beneficial for single-cell immune repertoire analysis, allowing researchers to study clonal diversity and specificity at the individual cell level.
Limitations: Although it provides long reads, Oxford Nanopore has lower accuracy compared to NGS and PacBio, which can affect the reliability of results in certain applications.
Applications of BCR and TCR Sequencing
BCR and TCR sequencing are pivotal tools in both research and clinical settings, offering profound insights into various aspects of immunology. Key applications of these methodologies include:
Cancer Immunotherapy: TCR sequencing facilitates the tracking of immune responses to tumors, thereby informing the development of more effective cancer therapies. A notable study by Valpione et al. demonstrated that baseline diversity of TCRs in tumor-infiltrating lymphocytes serves as a prognostic marker across various cancers. Moreover, TCR clonality prior to treatment has been identified as predictive of the efficacy of PD-1 blockade immunotherapy in metastatic melanoma.
Vaccine Development: BCR sequencing is instrumental in identifying antibodies targeting specific pathogens, while TCR sequencing enhances our understanding of the immune response to vaccines. Research by Rosati et al. explored the application of TCR sequencing to monitor vaccination responses, revealing detectable TCR expansion post-vaccination. However, further research is necessary to establish TCR expansion as a reliable biomarker for vaccine-induced protection.
Autoimmune Diseases: The sequencing of BCRs and TCRs aids in identifying clonal expansions linked to autoimmune conditions, potentially leading to improved diagnostic and therapeutic strategies. Zhang et al. employed single-cell RNA sequencing alongside TCR/BCR sequencing to profile immune cells and their repertoires in patients with systemic lupus erythematosus (SLE), thus providing valuable insights into the pathogenesis of the disease.
Infectious Diseases: Both BCR and TCR sequencing are crucial for studying immune responses to infections such as HIV, influenza, and COVID-19. Rosati et al. underscored the potential of single-cell TCR/BCR sequencing in elucidating immune responses to infectious diseases, enabling comprehensive analysis of immune cell heterogeneity and antigen-specific responses.
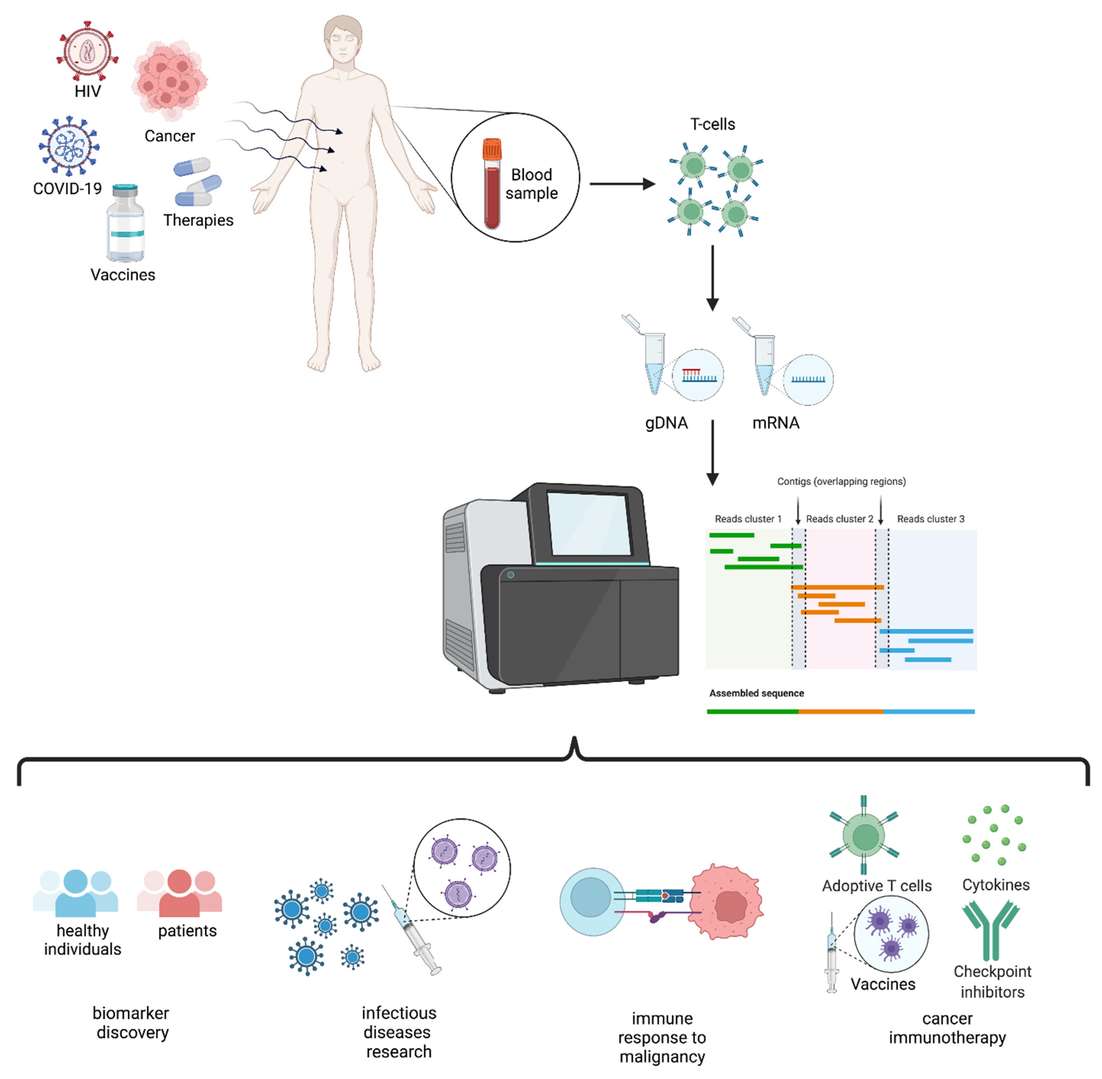 Figure 2. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
Figure 2. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
For more about applications of TCR sequencing, check out our TCR Detection Application and Method blog.
Comparison of BCR and TCR Repertoire Diversity
Here is a comparison between BCR and TCR sequencing in terms of diversity, sequencing depth, and challenges:
| Feature |
BCR Sequencing |
TCR Sequencing |
Scientific Example |
| Target |
Immunoglobulin genes |
T-cell receptor genes |
Briney et al. (2019) estimated 10^11 unique BCRs in an individual, demonstrating the exceptional diversity of the human antibody repertoire. |
| Diversity |
Higher (due to SHM) |
Lower |
Same as above |
| Sequencing Depth Required |
Higher |
Lower |
Bashford-Rogers et al. (2014) recommended 1,000,000 reads for comprehensive BCR analysis to capture rare clones. |
| Typical Usable Sequences |
55-76% |
60-76% |
Bashford-Rogers et al. (2018) reported 60-76% usable sequences for both BCR and TCR sequencing using PCR amplification methods. |
| Clonal Expansion Detection |
More challenging |
Easier |
Valpione et al. (2021) used TCR diversity in tumor-infiltrating lymphocytes as a prognostic marker in various cancers, highlighting easier detection of clonal expansions in TCR repertoires. |
| Cost |
Higher |
Lower |
Inferred from higher sequencing depth requirements for BCR sequencing. |
Technical Challenges and Solutions in BCR and TCR Sequencing
BCR and TCR analysis technologies have revolutionized our understanding of the immune system, but they face several technical challenges. These challenges include sequencing bias, accuracy issues, and difficulties in analyzing clonal diversity. Below, we explore these challenges, current solutions, and predict future developments.
Technical Challenges
Sequencing Bias:
Description: Sequencing bias can occur during PCR amplification when certain sequences are preferentially amplified over others due to differences in primer binding efficiency. This can lead to an inaccurate representation of the true diversity of the immune repertoire.
Impact: Bias affects the detection of rare clones and can skew results, leading to misinterpretations in studies related to immune responses.
Accuracy Issues:
Description: Errors introduced during sequencing can compromise the reliability of TCR and BCR data. These errors may arise from the sequencing process itself or from amplification biases.
Impact: Inaccurate sequences can hinder the identification of specific TCRs or BCRs that are critical for understanding immune functions and responses.
Clonal Diversity Analysis:
Description: Traditional bulk sequencing methods may not capture the full extent of clonal diversity, particularly in heterogeneous samples where rare clones are present at low frequencies.
Impact: This limitation restricts our ability to fully characterize immune responses, especially in contexts like cancer immunotherapy where understanding clonal dynamics is crucial.
Current Solutions
Single-Cell Sequencing Technologies:
Single-cell sequencing allows for the analysis of TCR and BCR repertoires at the individual cell level. This method enhances the detection of clonal diversity by enabling researchers to resolve clonal expansions and specific immune responses without the confounding effects of bulk population averaging.
Benefits: By capturing full-length sequences from individual cells, single-cell methods provide a clearer picture of clonal relationships and functional states within the immune system.
Unbiased Gene Amplification Technologies:
Techniques such as adaptor-ligation PCR have been developed to minimize PCR bias by ensuring even amplification across all TCR and BCR genes. This method uses a specific primer for the constant region combined with an adaptor primer, allowing for more uniform amplification.
Benefits: This approach reduces bias and improves accuracy in repertoire analysis, making it easier to capture a representative sample of the immune repertoire .
Error Correction Strategies:
The introduction of molecular barcodes has proven effective in correcting sequencing errors. Molecular barcodes tag individual molecules before amplification, allowing researchers to distinguish between true biological signals and artifacts introduced during PCR .
Additionally, advanced bioinformatics tools have been developed to correct errors by aligning sequences and identifying mismatches based on known rearrangement patterns .
Machine Learning for Data Processing:
Machine learning algorithms are increasingly being used to optimize data processing and interpretation in TCR/BCR sequencing. These algorithms can identify patterns in large datasets, correct biases, and enhance the sensitivity of rare clone detection.
Benefits: By leveraging machine learning, researchers can improve the accuracy of their analyses and gain deeper insights into clonal dynamics and immune responses .
Future Development
Integration of Technologies:
The future may see greater integration of single-cell sequencing with advanced bioinformatics and machine learning approaches. This could lead to comprehensive platforms that automate data processing from sample preparation to analysis, improving efficiency and reducing human error.
Enhanced Sensitivity and Accuracy:
Continued advancements in sequencing technologies will likely focus on increasing sensitivity for detecting rare clones while maintaining high accuracy. Innovations such as improved reagents and optimized protocols will be essential for achieving these goals.
Clinical Applications:
As TCR/BCR sequencing technologies mature, their applications in clinical settings will expand significantly. Personalized medicine approaches will benefit from precise immune repertoire profiling, aiding in treatment decisions for conditions like cancer and autoimmune diseases.
Standardization of Protocols:
The establishment of standardized protocols for TCR/BCR sequencing will be crucial for ensuring reproducibility across studies. This includes defining best practices for sample preparation, amplification methods, and data analysis techniques.
By addressing these technical challenges through innovative solutions, BCR and TCR sequencing technologies will continue to evolve, providing invaluable insights into immune function and paving the way for advancements in immunotherapy and personalized medicine.
Integrating BCR and TCR Data for Comprehensive Immune Profiling
The future landscape of immune profiling is poised to be transformed by the integration of BCR and TCR data, offering a comprehensive perspective on immune responses. This integrative approach is facilitated by cutting-edge technologies such as single-cell RNA sequencing (scRNA-seq) and NGS, which collectively enable the simultaneous analysis of BCR and TCR repertoires. Such advancements are pivotal for enhancing the monitoring of immune responses in contexts like cancer immunotherapy, autoimmune diseases, and various other immunological conditions.
Recent scientific research underscores the potential and impact of integrating BCR and TCR data:
Combined BCR and TCR Analysis in Multiple Sclerosis: Zhang et al. developed a sophisticated deep learning framework, scNAT, which seamlessly integrates paired scRNA-seq and scTCR-seq data. This methodology facilitated the identification of cell clusters and tracked T cell migration from blood to cerebrospinal fluid in multiple sclerosis patients, illustrating the utility of combined BCR and TCR analyses in deciphering complex immune responses.
Advanced BCR Sequencing Technology: Lau et al. described an innovative method for retrieving paired, full-length variable region sequences of BCRs from 3'-barcoded scRNA-seq libraries. This technique enables the concurrent analysis of BCR sequences and the entire transcriptomes of individual B cells, offering a more comprehensive understanding of B cell biology and antibody responses.
Integration with Transcriptomics: Wang et al. highlighted how the amalgamation of BCR and TCR sequencing with transcriptomics has been applied to diverse immunological conditions. This combined approach has been particularly insightful in exploring the tumor immune microenvironment, autoimmune diseases, infectious diseases, and chronic inflammatory conditions.
As technological frontiers expand, the integration of BCR and TCR sequencing promises to facilitate the development of personalized immunotherapies, thereby enhancing clinical outcomes. The synthesis of these datasets provides a deeper understanding of the immune system's intricacies, paving the way for more precise and potent therapeutic interventions.
Analysis Results between TCR Sequencing and BCR Sequencing
Methodological Comparisons
Reconstruction Accuracy:
A study utilizing the TRUST4 algorithm demonstrated significant differences in the reconstruction of TCR and BCR repertoires from bulk RNA-seq data. TRUST4 was able to call 281% more CDR3 sequences than MiXCR for TCRs, indicating its superior performance in capturing TCR diversity. In contrast, when evaluating BCRs, TRUST4 showed better precision (>18%) and sensitivity (>74%) compared to MiXCR across multiple tumor samples, suggesting that while both receptors present unique challenges, advancements in computational tools are improving accuracy for both types of sequencing.
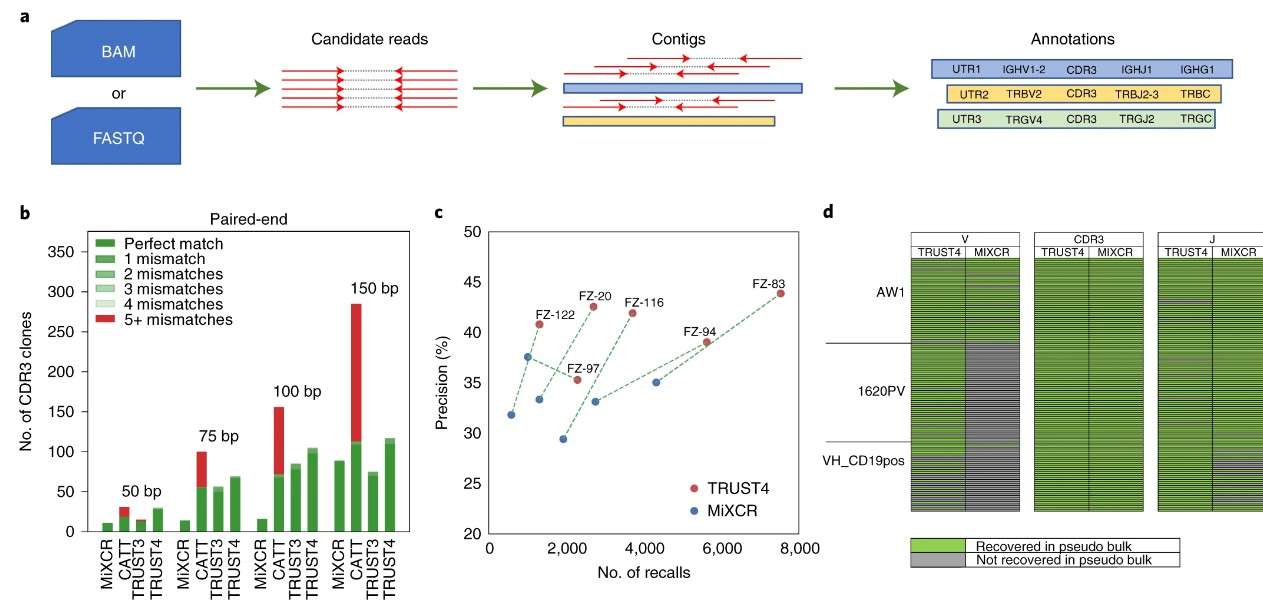 Figure 3: TRUST4 applied to bulk RNA-seq data.
Figure 3: TRUST4 applied to bulk RNA-seq data.
Sequencing Depth Requirements:
Research indicates that BCR sequencing often requires greater sequencing depth due to the high diversity generated by somatic hypermutation. For instance, a study found that achieving reliable data for BCRs necessitated up to 50 times higher sequencing depth compared to TCRs to capture equivalent numbers of specific reads. This difference underscores the complexities involved in analyzing BCR repertoires versus TCR repertoires.
Expression Levels:
The expression levels of TCRs and BCRs also influence analysis outcomes. For example, the TRUST4 analysis on single-cell RNA-seq data revealed that it recovered a higher percentage of BCR CDR3 sequences (78%) compared to TCR CDR3 sequences (48.1%). This discrepancy is attributed to the generally higher expression levels of BCRs in B cells compared to T cells.
Visualization of Results
Advanced analytical techniques allow for detailed visualization of TCR and BCR repertoire data. For example, 2D graphs can illustrate the usage percentages of V and J genes within the TCR and BCR repertoires, enabling comparisons across samples. Additionally, 3D visualizations can depict the frequency of VJ gene combinations, providing a comprehensive overview of clonal diversity and potential shifts in immune responses over time.
Clonal Architecture and Diversity
Recent explorations into the clonal architecture of BCRs and TCRs have shown that while both repertoires exhibit covarying patterns of clonality and diversity, their relationships can be altered under different immune conditions. Studies have utilized forest plots to display correlations between summary statistics from independent datasets, revealing how specific clonotypes may co-occur within individuals. This information is critical for understanding how immune responses are shaped by both B and T cell activities.
The analysis results from comparing TCR and BCR sequencing methodologies reveal significant differences in their reconstruction accuracy, sequencing depth requirements, expression levels, and clonal architecture. As computational tools continue to evolve, they enhance our ability to analyze these complex immune repertoires effectively. Understanding these distinctions not only aids in basic immunological research but also has profound implications for developing targeted therapies and improving patient outcomes in various diseases.
Why Choose CD Genomics for BCR and TCR Sequencing?
At CD Genomics, we are committed to advancing immunological research by providing expert immune repertoire and TCR sequencing services. Our offerings are distinguished by high-quality and reproducible results, tailored to the specific requirements of both researchers and clinicians. Leveraging cutting-edge technologies, we deliver customizable sequencing solutions ideal for exploring diverse areas such as immune responses to diseases, vaccine development, and cancer immunotherapy. Our comprehensive services encompass every stage of the sequencing process, from meticulous experimental design to sophisticated bioinformatics analysis, ensuring robust support for your research endeavors.
References:
-
Dong, D., Meng, F., Yang, C., Zhang, Y., Hao, Q., & Huang, Z. (2022). A structural platform for B cell receptor signaling. Cell Research, 32(11), 953-955. https://doi.org/10.1038/s41422-022-00724-9
-
Kovaltsuk, A., Leem, J., Kelm, S., Snowden, J., Deane, C. M., & Krawczyk, K. (2020). Observed Antibody Space: A Resource for Data Mining Next-Generation Sequencing of Antibody Repertoires. The Journal of Immunology, 204(9), 2502-2509. https://doi.org/10.4049/jimmunol.1901029
-
Rossjohn, J., Gras, S., Miles, J. J., Turner, S. J., Godfrey, D. I., & McCluskey, J. (2015). T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annual Review of Immunology, 33(1), 169-200. https://doi.org/10.1146/annurev-immunol-032414-112334
-
Newell, E. W., & Davis, M. M. (2014). Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nature Biotechnology, 32(2), 149-157. https://doi.org/10.1038/nbt.2783
-
Sela-Culang, I., Kunik, V., & Ofran, Y. (2013). The Structural Basis of Antibody-Antigen Recognition. Frontiers in Immunology, 4, 302. https://doi.org/10.3389/fimmu.2013.00302
-
Garcia, K. C., Adams, J. J., Feng, D., & Ely, L. K. (2012). The molecular basis of TCR germline bias for MHC is surprisingly simple. Nature Immunology, 13(5), 421-424. https://doi.org/10.1038/ni.2324
-
Murugan, A., Mora, T., Walczak, A. M., & Callan, C. G. (2012). Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proceedings of the National Academy of Sciences, 109(40), 16161-16166. https://doi.org/10.1073/pnas.1212755109
-
Elhanati, Y., Sethna, Z., Marcou, Q., Callan, C. G., Mora, T., & Walczak, A. M. (2015). Inferring processes underlying B-cell repertoire diversity. Philosophical Transactions of the Royal Society B: Biological Sciences, 370(1676), 20140243. https://doi.org/10.1098/rstb.2014.0243
-
Victora, G. D., & Nussenzweig, M. C. (2012). Germinal Centers. Annual Review of Immunology, 30(1), 429-457. https://doi.org/10.1146/annurev-immunol-020711-075032
-
Qi, Q., Liu, Y., Cheng, Y., Glanville, J., Zhang, D., Lee, J. Y., Olshen, R. A., Weyand, C. M., Boyd, S. D., & Goronzy, J. J. (2014). Diversity and clonal selection in the human T-cell repertoire. Proceedings of the National Academy of Sciences, 111(36), 13139-13144. https://doi.org/10.1073/pnas.1409155111
-
Valpione, S., Mundra, P. A., Galvani, E., Campana, L. G., Lorigan, P., De Rosa, F., Gupta, A., Middleton, M. R., & Lorigan, P. (2021). The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nature Communications, 12(1), 4098. https://doi.org/10.1038/s41467-021-24343-x
-
Rosati, E., Pogorelyy, M. V., Minervina, A. A., Franke, A., Bains, I., Hickman, S. P., Chudakov, D. M., & Mora, T. (2023). On the feasibility of using TCR sequencing to follow a vaccination response. Frontiers in Immunology, 14, 1210168. https://doi.org/10.3389/fimmu.2023.1210168
-
Zhang, F., Wei, K., Slowikowski, K., Fonseka, C. Y., Rao, D. A., Kelly, S., Goodman, S. M., Tabechian, D., Hughes, L. B., Salomon-Escoto, K., Watts, G. F. M., Jonsson, A. H., Rangel-Moreno, J., Meednu, N., Rozo, C., Apruzzese, W., Eisenhaure, T. M., Lieb, D. J., Boyle, D. L., ... Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium. (2019). Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nature Immunology, 20(7), 928-942. https://doi.org/10.1038/s41590-019-0378-1
-
Rosati, E., Dowds, C. M., Liaskou, E., Henriksen, E. K. K., Karlsen, T. H., & Franke, A. (2017). Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnology, 17(1), 61. https://doi.org/10.1186/s12896-017-0379-9
-
Briney, B., Inderbitzin, A., Joyce, C., & Burton, D. R. (2019). Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature, 566(7744), 393-397. https://doi.org/10.1038/s41586-019-0879-y
-
Bashford-Rogers, R. J. M., Palser, A. L., Huntly, B. J., Rance, R., Vassiliou, G. S., Follows, G. A., & Kellam, P. (2014). Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Research, 24(11), 1734-1744. https://doi.org/10.1101/gr.154815.113
-
Bashford-Rogers, R. J. M., Smith, K. G. C., & Thomas, D. C. (2018). Antibody repertoire analysis in polygenic autoimmune diseases. Immunology, 155(1), 3-17. https://doi.org/10.1111/imm.12927
-
Zhang, Y., Hu, Y., Bian, X., Wang, Y., Wang, K., Xue, Z., & Jiang, T. (2023). scNAT: a deep learning method for integrating paired single-cell RNA and T cell receptor sequencing profiles. Genome Biology, 24, 292. https://doi.org/10.1186/s13059-023-03129-y
-
Lau, A. T. Y., Nguyen, L. T., Fink, K., MacIsaac, J. L., Curtis, J. E., Marra, M. A., & Holt, R. A. (2024). Full-length single-cell BCR sequencing paired with RNA expression from 3'-barcoded libraries. Communications Biology, 7(1), 225. https://doi.org/10.1038/s42003-024-06823-0
-
Wang, X., Wen, Y., Xue, J., & Lu, Q. (2022). Research progress on application of single-cell TCR/BCR sequencing in immune system. Frontiers in Immunology, 13, 969808. https://doi.org/10.3389/fimmu.2022.969808
-
Zhang, Y., et al. (2021). TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nature Biotechnology, 39(5), 703-711. https://doi.org/10.1038/s41592-021-01142-2
-
O'Connor, R.S., et al. (2014). A comparison of B-cell receptor sequencing methods: implications for immunotherapy development. Nature Communications, 5(1), 1-10. https://doi.org/10.1038/ncomms4243
-
Repertoire Genesis, Inc. (2023). TCR/BCR Repertoire Analysis: Our Technologies. Retrieved from https://www.repertoire.co.jp/en/research/technology/repertoire/
-
Madi, A., et al. (2022). Exploration of shared features of B cell receptor and T cell receptor repertoires in human health and disease. Nature Communications, 13(1), 1-13. https://doi.org/10.1038/s41467-022-33948-5


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1: Approaches for BCR and TCR repertoire sequencing. (Yu Fen Samantha Seah et al,. 2017)
Figure 1: Approaches for BCR and TCR repertoire sequencing. (Yu Fen Samantha Seah et al,. 2017) Figure 2. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022)
Figure 2. Schematic representation of TCR repertoire analysis and applications. (Lucia Mazzotti et al,. 2022) Figure 3: TRUST4 applied to bulk RNA-seq data.
Figure 3: TRUST4 applied to bulk RNA-seq data.