PCR, a widely used gene cloning technique, enables efficient in vitro replication of specific DNA fragments. Utilizing PCR technology, we can amplify target genes to sufficient quantities, meeting the demands of subsequent sequencing and analysis.
In current research, there are typically two main methods for sequence analysis of PCR products. Firstly, direct Sanger sequencing is employed, suitable for detecting gene mutations with clear loci, such as limited single-gene hereditary diseases and microbial population identification, HLA typing detection (PCR-SBT), among others. Secondly, PCR products are subjected to vector construction, commonly employing T vectors, followed by transformation into Escherichia coli. Subsequent steps involve culture, colony picking, and single clone sequencing with in-depth analysis.
The application scope of direct PCR product sequencing technology is extensive. It is not only used for gene mutation detection but also in microbial population identification, HLA typing detection, and validating NGS results, providing robust support for scientific research endeavors.
You may interested in
Learn More
Application of Sanger Sequencing in PCR Product Analysis
Mutation Detection
Mutational events induce variability in DNA sequences. Sanger sequencing, as an efficient and precise detection method, effectively identifies these mutations, including single nucleotide polymorphisms (SNPs), insertions, and deletions. Mutation detection plays an indispensable role in crucial research domains such as hereditary diseases and cancer.
The basic process of mutation detection is outlined as follows:
Firstly, specific primers are designed for target exonic sequences prone to mutations, ensuring accurate capture of the target region. Subsequently, DNA is extracted from the sample to serve as the foundation material for subsequent experiments.
Next, target DNA sequences are amplified using PCR amplification techniques, and the amplification products are purified to eliminate the influence of nonspecific amplification and impurities. Following purification of the amplification products, their sizes are determined through methods such as gel electrophoresis to ensure that the products meet expectations.
Subsequently, Sanger sequencing of the amplification products is conducted to obtain DNA sequence information from the target region. Upon completion of sequencing, the obtained sequence chromatogram is compared and analyzed with the reference sequence of the target gene to identify existing mutation sites and their types.
Finally, to ensure the accuracy and reliability of the sequencing results, optional validation steps can be undertaken post-sequencing. This can be achieved through methods such as fluorescent quantification detection kits (using the ARMS method with a sensitivity of 1%), further validating the accuracy of mutation detection results.
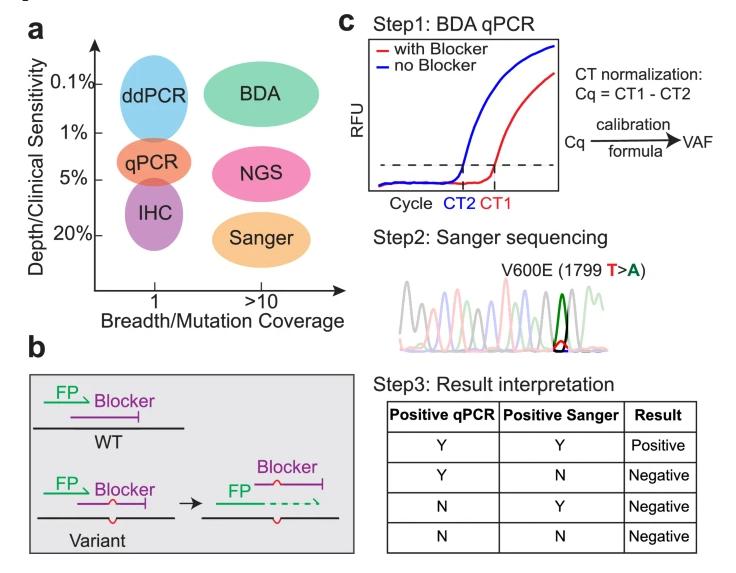 Methods for the Detection of BRAF Mutations. (a) Comparative Assessment of Depth and Breadth in BRAF Mutation Detection Methods. (b) Mechanism for Blocker-Displacement Amplification (BDA) Variant Enrichment. The Blocker selectively binds to and inhibits the PCR amplification of wildtype (WT) DNA sequences, facilitating the preferential amplification of BRAF mutations. (c) Workflow of the BDA Mutation Detection Assay. The BDA assay is initially conducted in quantitative PCR (qPCR), followed by Sanger sequencing of the amplicons to pinpoint the specific mutation.
Methods for the Detection of BRAF Mutations. (a) Comparative Assessment of Depth and Breadth in BRAF Mutation Detection Methods. (b) Mechanism for Blocker-Displacement Amplification (BDA) Variant Enrichment. The Blocker selectively binds to and inhibits the PCR amplification of wildtype (WT) DNA sequences, facilitating the preferential amplification of BRAF mutations. (c) Workflow of the BDA Mutation Detection Assay. The BDA assay is initially conducted in quantitative PCR (qPCR), followed by Sanger sequencing of the amplicons to pinpoint the specific mutation.
Microbes Identification
Microbes identification endeavors to achieve precise characterization and classification of diverse microorganisms encompassing bacteria, fungi, viruses, and parasites utilizing Sanger sequencing. Presented below is a concise delineation of the identification protocol:
Initially, meticulous design of specific primers targets representative conserved regions within the microbial DNA of interest, such as the 16S rDNA for bacteria or the ITS region for fungi.
Subsequent to primer design, efficient amplification of the conserved regions is executed employing PCR technology to yield an ample supply of DNA fragments requisite for downstream analysis.
Following successful amplification, first-generation sequencing of the PCR products ensues to discern the DNA sequences accurately, thereby elucidating the genetic makeup of the target microorganisms.
Upon procurement of sequencing outcomes, rigorous sequence alignment and analysis against globally acknowledged gene repositories like NCBI GenBank are conducted. This meticulous alignment facilitates the determination of microbial species relationships.
The entirety of the identification process is characterized by its rigorous and methodical nature, ensuring the precision and dependability of the identification outcomes, thus furnishing robust support for microbial research and practical applications.
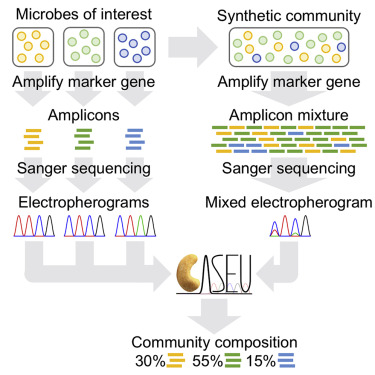 Quantify bacterial communities through Sanger sequencing. Here, the authors quantify amplicon composition in synthetic bacterial communities through Sanger sequencing. They PCR amplify a universal marker gene, and subsequently, they sequence this amplicon mixture in a single Sanger sequencing reaction.
Quantify bacterial communities through Sanger sequencing. Here, the authors quantify amplicon composition in synthetic bacterial communities through Sanger sequencing. They PCR amplify a universal marker gene, and subsequently, they sequence this amplicon mixture in a single Sanger sequencing reaction.
HLA Typing
Compared to PCR-based methods such as PCR-sequence-specific oligonucleotide probe assays (low resolution) and PCR-sequence-specific primer gene amplification (medium resolution), PCR-based direct sequencing typing (sequence-based typing, SBT) offers higher resolution. Among these techniques, classical Sanger sequencing serves as an optional sequencing approach for PCR-based direct sequencing typing.
The workflow is outlined as follows: Initially, site-specific primers are utilized to selectively amplify critical HLA loci, ensuring the precise acquisition of target sequences. Subsequently, the amplified products undergo Sanger sequencing to obtain comprehensive base sequence information. Finally, the sequencing results are thoroughly analyzed using software to acquire high-resolution HLA gene typing information. Throughout this process, the sequencing results are aligned with known HLA databases to accurately determine an individual's specific HLA genotype, aiding in the discovery of novel allelic variations.
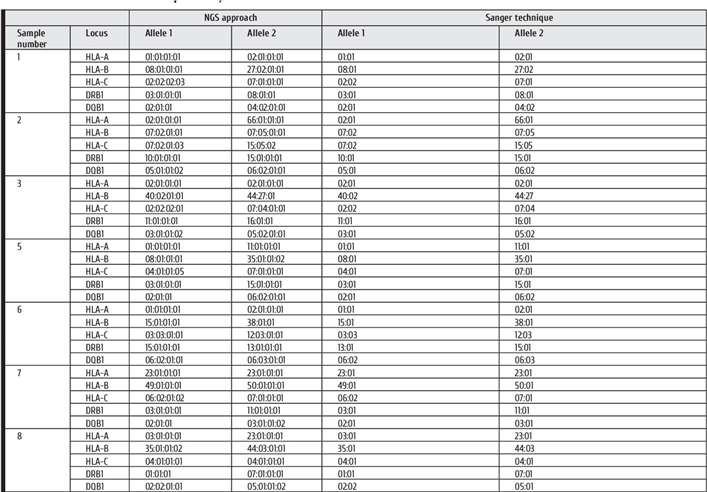 Comparative analysis of NGS and Sanger sequencing methods for HLA typing
Comparative analysis of NGS and Sanger sequencing methods for HLA typing
NGS Results Validation
Given the multifaceted uncertainties affecting the accuracy of NGS, stemming from variables such as targeted enrichment systems, sequencing platforms, bioinformatics pipelines, and variant interpretation expertise, reliable validation tools are indispensable for comparative verification. Among these, Sanger sequencing technology, renowned for its capability to sequence a single strand of DNA segment at a time with an accuracy reaching 99.99%, is widely acknowledged and consistently employed as the gold standard for validation.
The validation process is summarized as follows:
Initially, precise primer design targeting the identified variant loci from NGS detection ensures the accurate capture of the target regions. Subsequently, through amplification of the target segments and purification of the products, high-quality sequencing templates are obtained. Next, Sanger sequencing technology is utilized to sequence the processed templates, yielding detailed sequence information. Finally, thorough analysis of the Sanger sequencing results regarding variant loci confirms their consistency with the NGS detection outcomes. Through this rigorous validation process, we can more accurately assess the reliability of NGS results, providing robust support for subsequent scientific research or clinical decision-making.
Mitochondrial DNA Sequencing
Mitochondrial DNA sequencing is a precise analytical technique targeting specific loci. In cases of suspected mitochondrial gene mutations, peripheral venous blood samples are first collected from the patients, from which genomic DNA is extracted. Subsequently, advanced NGS technology comprehensively examines the patient's entire mitochondrial genome sequence. Based on these preliminary detection results, further validation analysis of the patient's mitochondrial DNA is performed using the Sanger sequencing method on samples obtained from the patient's parents and siblings to ensure the accuracy and reliability of the results. Through this series of rigorous sequencing processes, robust molecular genetic evidence is provided for clinical diagnosis and treatment.
Given the esteemed reputation of Sanger sequencing for its outstanding high-precision characteristics, its core detection principle lies in the ability to accurately read DNA sequences base by base. Therefore, meticulous control of the quality of PCR products, including their concentration and purity, is crucial for the success of Sanger sequencing.
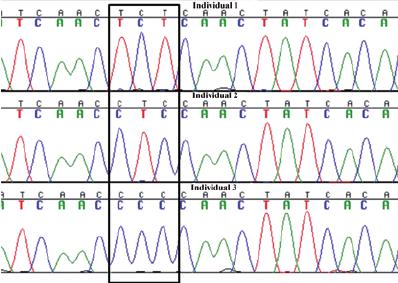 Sanger sequencing of mtDNA. Electropherograms generated by Sanger dideoxy sequencing of mtDNA extracted from three ancient bone remains found in a grave from the 10 th century in Sigtuna, mid Sweden. (Martina Nilsson)
Sanger sequencing of mtDNA. Electropherograms generated by Sanger dideoxy sequencing of mtDNA extracted from three ancient bone remains found in a grave from the 10 th century in Sigtuna, mid Sweden. (Martina Nilsson)
Optimizing Sanger Sequencing: Troubleshooting Common Issues
During the preparatory phase of Sanger sequencing, continuous optimization of experimental conditions is necessary to maximize sequence quality and accuracy. This involves, among other aspects, fine-tuning the design of PCR primers and precisely adjusting PCR reaction conditions. Additionally, the purification of products after PCR reaction is indispensable. This step aims to effectively remove impurities such as nonspecifically amplified fragments and primer sequences, thereby ensuring the accuracy of the target gene sequence.
However, in practical operation, factors such as the concentration, purity, and structure of the PCR products to be sequenced may lead to a series of suboptimal sequencing results. To address this, we have compiled a list of common issues and corresponding troubleshooting strategies for practical reference.
| Sequencing Peak Pattern |
Possible Causes |
Recommended Solutions |
| Weak Signal |
Low concentration of PCR product |
Sequence as soon as possible after amplification (or perform secondary amplification) |
| Signal Decay |
Presence of special nucleic acid structures (e.g., repeat sequences, GC-rich regions, AT-rich regions) |
Perform reverse sequencing from the other end of the sample, and splice to obtain the complete sequence. |
| Signal Interruption |
- |
- |
| Local Frameshift |
- |
- |
| Peak Overlay |
Double Peaks |
- Non-specific amplification: Adjust PCR amplification system and re-amplify.
- Front-end double peaks: PCR products contain multiple primer binding sites, consider changing sequencing primers.
- PCR products contain primer dimers or small fragment contamination: Gel purification of PCR products. |
|
|
- Middle double peaks: Some products have base deletion, clone PCR products for sequencing.
- PCR products are impure due to gene alleles, clone PCR products for sequencing.
- Rear-end double peaks: Some products have base deletion, clone PCR products for sequencing. |
References:
- Cheng, L.Y., Haydu, L.E., Song, P. et al. High sensitivity sanger sequencing detection of BRAF mutations in metastatic melanoma FFPE tissue specimens. Sci Rep 11, 9043 (2021).
- Cermak N, Datta MS, Conwill A. Rapid, Inexpensive Measurement of Synthetic Bacterial Community Composition by Sanger Sequencing of Amplicon Mixtures. iScience. 2020
- Syndercombe Court D. Mitochondrial DNA in forensic use. Emerg Top Life Sci. 2021


 Sample Submission Guidelines
Sample Submission Guidelines
 Methods for the Detection of BRAF Mutations. (a) Comparative Assessment of Depth and Breadth in BRAF Mutation Detection Methods. (b) Mechanism for Blocker-Displacement Amplification (BDA) Variant Enrichment. The Blocker selectively binds to and inhibits the PCR amplification of wildtype (WT) DNA sequences, facilitating the preferential amplification of BRAF mutations. (c) Workflow of the BDA Mutation Detection Assay. The BDA assay is initially conducted in quantitative PCR (qPCR), followed by Sanger sequencing of the amplicons to pinpoint the specific mutation.
Methods for the Detection of BRAF Mutations. (a) Comparative Assessment of Depth and Breadth in BRAF Mutation Detection Methods. (b) Mechanism for Blocker-Displacement Amplification (BDA) Variant Enrichment. The Blocker selectively binds to and inhibits the PCR amplification of wildtype (WT) DNA sequences, facilitating the preferential amplification of BRAF mutations. (c) Workflow of the BDA Mutation Detection Assay. The BDA assay is initially conducted in quantitative PCR (qPCR), followed by Sanger sequencing of the amplicons to pinpoint the specific mutation. Quantify bacterial communities through Sanger sequencing. Here, the authors quantify amplicon composition in synthetic bacterial communities through Sanger sequencing. They PCR amplify a universal marker gene, and subsequently, they sequence this amplicon mixture in a single Sanger sequencing reaction.
Quantify bacterial communities through Sanger sequencing. Here, the authors quantify amplicon composition in synthetic bacterial communities through Sanger sequencing. They PCR amplify a universal marker gene, and subsequently, they sequence this amplicon mixture in a single Sanger sequencing reaction. Comparative analysis of NGS and Sanger sequencing methods for HLA typing
Comparative analysis of NGS and Sanger sequencing methods for HLA typing Sanger sequencing of mtDNA. Electropherograms generated by Sanger dideoxy sequencing of mtDNA extracted from three ancient bone remains found in a grave from the 10 th century in Sigtuna, mid Sweden. (Martina Nilsson)
Sanger sequencing of mtDNA. Electropherograms generated by Sanger dideoxy sequencing of mtDNA extracted from three ancient bone remains found in a grave from the 10 th century in Sigtuna, mid Sweden. (Martina Nilsson)