What is Microbial Identification
Microbial identification entails the rigorous process of differentiating among various categories of microorganisms—encompassing bacteria, yeasts, and molds—and ascertaining their genus, species, or strain using established classification frameworks. This process involves a detailed characterization of unknown microorganisms based on an analysis of both phenotypic and genotypic traits.
Despite their microscopic size, microorganisms play a pivotal role within ecosystems. They significantly contribute to biodiversity in various environments including soil, water, and air. Additionally, they have profound implications for human health, participating in beneficial processes such as digestion and immune system regulation, while also being implicated in adverse outcomes including infections and diseases. As such, the detection, study, and identification of these microorganisms remain imperative for advancing our understanding of their roles and impacts.
Methods of Microbial Identification
Traditional Methods
Microbial identification traditionally hinges on phenotypic characteristics, including cell morphology, biochemical reactions, and specific growth requirements. Specifically, these methods encompass:
- Fatty Acid Analysis: This method identifies microorganisms based on their unique fatty acid profiles.
- Carbon Source Utilization: Here, the ability of microorganisms to metabolize various carbon sources is assessed.
- (G+C) Mol% Determination: This technique classifies microbes by evaluating the guanine-cytosine content in their DNA.
- API Analysis: Standardized biochemical assays are used to determine microbial species.
Though effective, these traditional methods can be labor-intensive and time-consuming, necessitating extensive manual effort.
Rapid Identification Techniques
With advancements in technology, rapid identification techniques offer more efficient and precise microbial identification. These methods provide quicker turnaround times and enhanced accuracy. Key techniques include:
- DNA-DNA Hybridization: Offering superior accuracy compared to (G+C) mol% determination, DNA-DNA hybridization relies on the principle that when DNA-DNA hybridization homology exceeds 70%, the species can be precisely identified.
- 16S rRNA Gene Sequencing: This methodology serves as a powerful tool for bacterial identification. By analyzing the 16S rRNA gene sequence in microbial samples and comparing it with a verified genetic sequencing library, CD Genomics can determine bacterial identities at both the genus and species levels, providing rapid and dependable results.
- 18S/ITS Sequencing: For the identification of fungi, particularly yeasts and molds, 18S/ITS sequencing is employed. This technique is both efficient and precise for identifying fungi at the genus and species levels.
- Whole Genome Sequencing (WGS): WGS provides detailed genetic analysis by sequencing the entire genome of a microorganism, aiding in distinguishing closely related species and understanding genetic variations.
- Metagenomics: Metagenomics involves sequencing genetic material directly from environmental samples, enabling the study of microbial communities, including those that cannot be cultured.
- MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry): This advanced method enables the reliable identification of bacteria and yeasts. Compared to traditional phenotypic methods, MALDI-TOF MS offers higher accuracy and faster turnaround times.
Our Microbial Identification Services
CD Genomics provides the following advanced microbial identification services:
- Sanger Sequencing: Employs rRNA gene sequences for accurate microbial identification, leveraging an extensive database to match sequences and identify bacteria and fungi precisely.
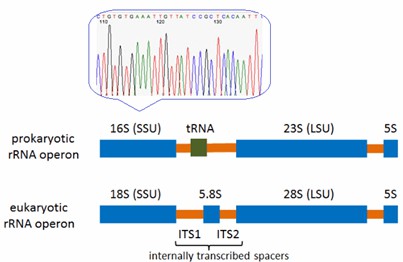
- Metagenomics: Sequences genetic material directly from environmental samples, enabling the study of diverse and unculturable microbial communities and providing a comprehensive view of microbial ecosystems.
- Microbial Whole Genome Sequencing: Analyzes the entire genome of microorganisms to distinguish closely related species and resolve classification ambiguities through detailed genetic profiling.
- 16S/ITS/18S Amplicon Sequencing: Utilizes next-generation sequencing technologies, including PacBio's long-read sequencing, for high-resolution analysis of microbial diversity and strain-level identification, surpassing traditional methods.
Selection of Sequencing Platforms and Protocols
In general, the accuracy of species identification improves with longer sequencing reads. However, practical considerations such as sequencing platform capabilities and costs often necessitate a choice of sequencing platform and amplification regions based on the specific objectives of the study.
- Traditional first-generation sequencing methods are noted for their high accuracy, relatively low cost, and long read lengths, which can nearly cover the entire length of the 16S rRNA gene. However, these methods are limited by their inability to directly analyze mixed or complex samples, resulting in lower sequencing throughput.
- Second-generation sequencing technologies offer high throughput, reduced costs, and the ability to perform library preparation and sequencing on mixed samples. This makes them a predominant method in 16S amplicon sequencing research. Nonetheless, these technologies are constrained by shorter read lengths, typically necessitating the amplification of specific regions such as V1–V3, V3–V4, or V4–V5, followed by paired-end sequencing (PE250 or PE300).
- The latest third-generation sequencing technologies provide ultra-long read lengths and high throughput, facilitating the acquisition of full-length 16S rRNA gene sequences with ease. Despite these advantages, they are hampered by higher error rates and costs, which currently limits their widespread application. However, with ongoing advancements and refinements, third-generation sequencing technologies are poised to become a mainstream method in microbial research in the future.
Advantages of Our Microbial Identification Services
- High-Throughput and Accuracy: CD Genomics leverages cutting-edge high-throughput sequencing platforms, notably the Illumina NovaSeq 6000, to ensure precise and dependable sequencing outcomes. Our advanced equipment is capable of processing large sample volumes efficiently, thus facilitating expedited turnaround times.
- Comprehensive Microbial Identification: Our suite of services encompasses the identification of a broad spectrum of microorganisms, including bacteria, fungi, and archaea. Our extensive genomic sequence database enables the accurate identification of most microbial species, thereby enhancing the reliability of our results.
- Robust Quality Management: We strictly adhere to a rigorous quality management system that underpins the accuracy and dependability of every test result. Automation within our processes mitigates manual errors and bolsters consistency across analyses.
Application of Microbial Identification
- Establishing Microbial Libraries: The accurate identification of microorganisms is fundamental for the creation and maintenance of microbial libraries. These repositories serve as essential resources for scientific research, permitting the study and comparison of diverse microbial species.
- Screening for Functional Microbial Communities: Identifying microorganisms that exhibit specific biological functions—such as enzyme production, acid production, or antibiotic synthesis—is critical for numerous industrial applications. Such functional screening allows for the targeted utilization of microbial communities to achieve desired outcomes.
- Microbial Product Development: In the context of microbial product development, such as in the production of probiotics or biofertilizers, the precise identification of beneficial microorganisms is paramount. This accuracy ensures both the efficacy and safety of the final products, facilitating their successful application.
- Environmental Monitoring: Microbial identification is also a pivotal component of environmental monitoring efforts, aiding in the detection and tracking of microbial contamination across water, soil, and air samples. This information is indispensable for maintaining environmental health and safeguarding public safety.
- Food and Pharmaceutical Quality Control: In the realms of food and pharmaceutical production, ensuring the absence of harmful microorganisms is of utmost importance. The rigorous identification of microbial contaminants aids in monitoring and controlling potential contamination, thereby ensuring product safety and regulatory compliance.
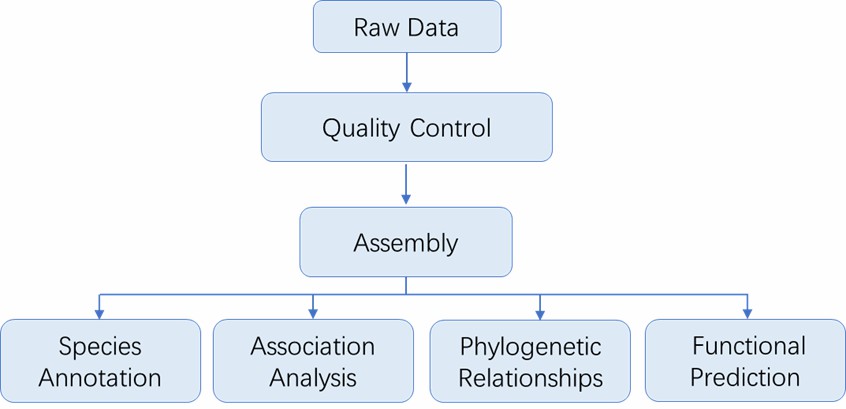
Microbial Identification Workflow
The workflow of microbial identification at CD Genomics involves several key steps:

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Microbial Identification for your writing (customization)
Partial results are shown below:

The taxonomy distribution of all sample in Phylum classification level.

Species abundance Heatmap.

Rarefaction curve of the sequenced reads for samples (The above figure) & The depth of the sequencing samples (The below figure).

Boxplot analysis based on bray Curtis (A), binary jaccard (B), unweighted unifrac (C), and weighted unifrac (D).

PCoA analysis based on bray Curtis (A), binary jaccard (B), unweighted unifrac (C), and weighted unifrac (D).

UPGMA clustering tree.

Mean proportion of treated and control group.

Cladogram.

LDA SCORE.
1. How to Select the Appropriate Sequencing Method?
Choosing the correct sequencing method depends on the research goals and sample types involved. For instance, Sanger sequencing is suited for pure cultured strains, metagenomic sequencing is ideal for studying the diversity of complex environmental samples, while whole-genome sequencing offers detailed genomic information to differentiate closely related species.
2. How are Results Presented?
Results are typically delivered in a comprehensive report format, which includes species annotations, phylogenetic trees, functional prediction outcomes, and various data visualizations. The report may feature statistics on sequence splits, bar charts of annotation levels, LEfSe analysis LDA bar charts, Venn diagrams, heatmaps of beta diversity indices, and species interaction networks.
3. How do I prepare my samples for submission?
Ensure that samples are properly labeled and stored according to the guidelines provided. For environmental samples, please use sterile containers and preserve samples to prevent contamination. Detailed instructions are available on our service pages.
4. Why is accurate bacterial identification essential?
Accurate bacterial identification is paramount for environmental monitoring (EM) programs within pharmaceutical and other regulated product manufacturing industries. The process of identifying unknown isolates serves as a crucial initial step in assessing the potential risks posed by microorganisms to the manufacturing environment, final product, and ultimately to patients. Effective bacterial identification services, which offer EM data tracking and trending solutions, are vital for maintaining a clear baseline understanding of a facility's microbial flora. This allows for the early detection of any unusual microbial activity, thereby providing a window of opportunity for timely remediation.
Tapping into the maize root microbiome to identify bacteria that promote growth under chilling conditions
Journal: Microbiome
Impact factor: 16.837
Published: 18 April 2020
Background
Maize faces yield issues in colder climates due to chilling temperatures. To address this, researchers investigated how chilling stress affects the maize root microbiome and identified beneficial bacteria that could enhance growth under stress. They analyzed the root microbiome using deep-sequencing technologies and screened a collection of maize endophytes for their growth-promoting effects under chilling conditions.
Materials & Methods
Sample Preparation
- Maize plants
- Root endosphere samples
- DNA extraction
Sequencing
- 16S rRNA sequencing
- Illumina MiSeq
- Demultiplexing and primer removal
- Quality check
- Asv determination
- Taxonomy assignment
- Statistical analysis
Results
Two experiments identified key bacterial communities in maize root endosphere grown in field soil and pots. Both conditions showed distinct microbiomes, with Proteobacteria, Bacteroidetes, Chloroflexi, Firmicutes, and Actinobacteria being predominant. Differences in abundance, notably in Actinobacteria and Oxalobacteraceae, were observed between field and pot-grown maize. The study highlights how growth conditions affect root microbiome composition.
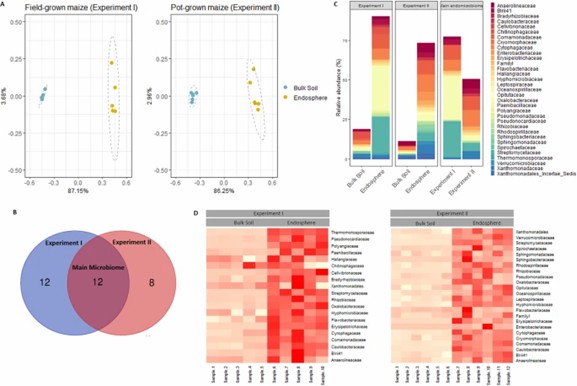 Fig 1. Identification of the main maize root endosphere families of field-grown and pot-grown maize.
Fig 1. Identification of the main maize root endosphere families of field-grown and pot-grown maize.
Chilling temperatures significantly alter the maize root endosphere microbiome, with over 40% of the variance attributed to temperature changes. The impact is more pronounced in the root endosphere than in the bulk soil, where specific bacterial families like Chitinophagaceae and Blastocatellaceae show notable shifts.
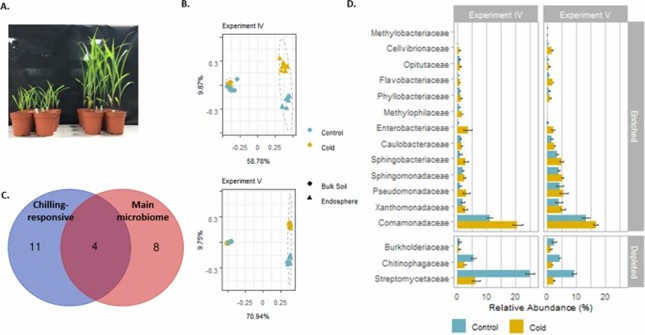 Fig 2. Bacterial community shifts upon chilling temperature treatment in experiments IV and V.
Fig 2. Bacterial community shifts upon chilling temperature treatment in experiments IV and V.
Conclusion
Plants establish a stable root microbiome primarily from surrounding soil, with chilling conditions causing more significant bacterial shifts in the roots than in the bulk soil. The identified PGPR strains, which thrive in the root endosphere, show promise for promoting maize growth under chilling temperatures and will be further explored for agricultural use.
Reference
- Beirinckx S, Viaene T, Haegeman A, et al. Tapping into the maize root microbiome to identify bacteria that promote growth under chilling conditions. Microbiome, 2020, 8: 1-13.
Here are some publications that have been successfully published using our services or other related services:
Transferrable protection by gut microbes against STING-associated lung disease
Journal: Cell Reports
Year: 2021
Microbial adaptation and response to high ammonia concentrations and precipitates during anaerobic digestion under psychrophilic and mesophilic conditions
Journal: Water Research
Year: 2021
Algal-bacterial synergy in treatment of winery wastewater
Journal: NPJ Clean Water
Year: 2018
Blocking IL-17A prevents oxycodone-induced depression-like effects and elevation of IL-6 levels in the ventral tegmental area and reduces oxycodone-derived physical dependence in rats
Journal: Brain, Behavior, and Immunity
Year: 2024
Indole-3-Propionic Acid, a Gut Microbiota Metabolite, Protects Against the Development of Postoperative Delirium
Journal: Annals of Surgery
Year: 2023
Elucidating the effects of organic vs. conventional cropping practice and rhizobia inoculation on rhizosphere microbial diversity and yield of peanut
Journal: Environmental Microbiome
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
